Protein sorting - 6.1
1/25
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
26 Terms
Protein Sorting
The process by which newly-made proteins are directed to the correct location
Each protein has a sorting signal (signal sequence) that can range from 3-60 continuous AAs. The signal is often but not always, and is removed once the protein arrives at its destination
It’s done in 3 major steps : recognition of the signal sequence by a shuttling cytosolic receptor; targeting to the outer surface of organelle membrane; import of targeted protein into membrane/ across membrane
How are proteins transported across a membrane that is normally impermeable to hydrophilic molecules?
Transport through nuclear pores : Nuclear pores transport specific proteins that must remain folded
Transport across membranes : across ER, mitochondria, chloroplast, peroxisome membranes → this requires protein translocators. In order to cross the membrane, proteins are unfolded
i.e. UPR that gets proteins degraded by proteasome in cytoplasm
Transport by vesicles : from ER onward & through endomembrane system → vesicles collect cargo protein & pinch off from membrane. Vesicles deliver their cargo by fusing with another compartment. Proteins remain folded during transport
NPC
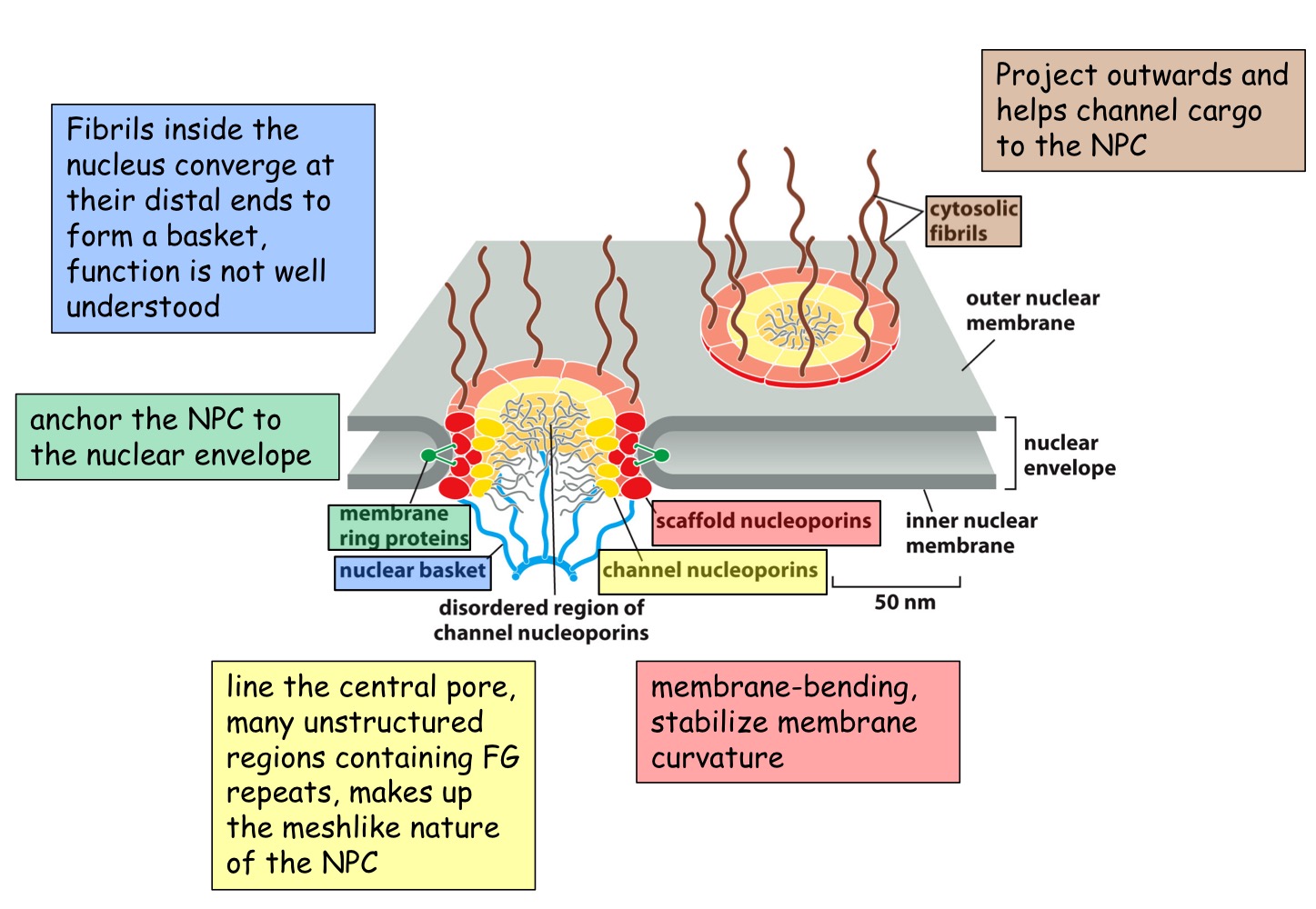
Nuclear Pore Complex
Huge complex (x15-30 mass of ribosome)
channel 20 to 30mm wide
FG (phenylannine-glycine) domains form a hydrophobic sieve that blocks diffusion of larger macromolecules (<40000 daltons)
Protein transport through an NPC
Proteins synthesized in the cytoplasm are targeted for the nucleus by a nuclear localization signal (NLS), e.g. P-K-K-K-R-K-V having basic residues
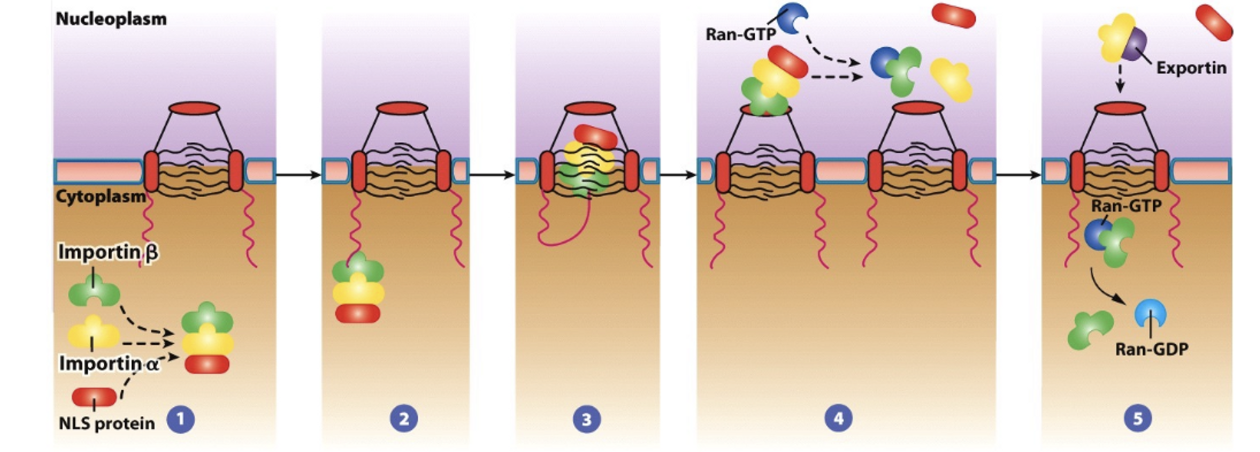
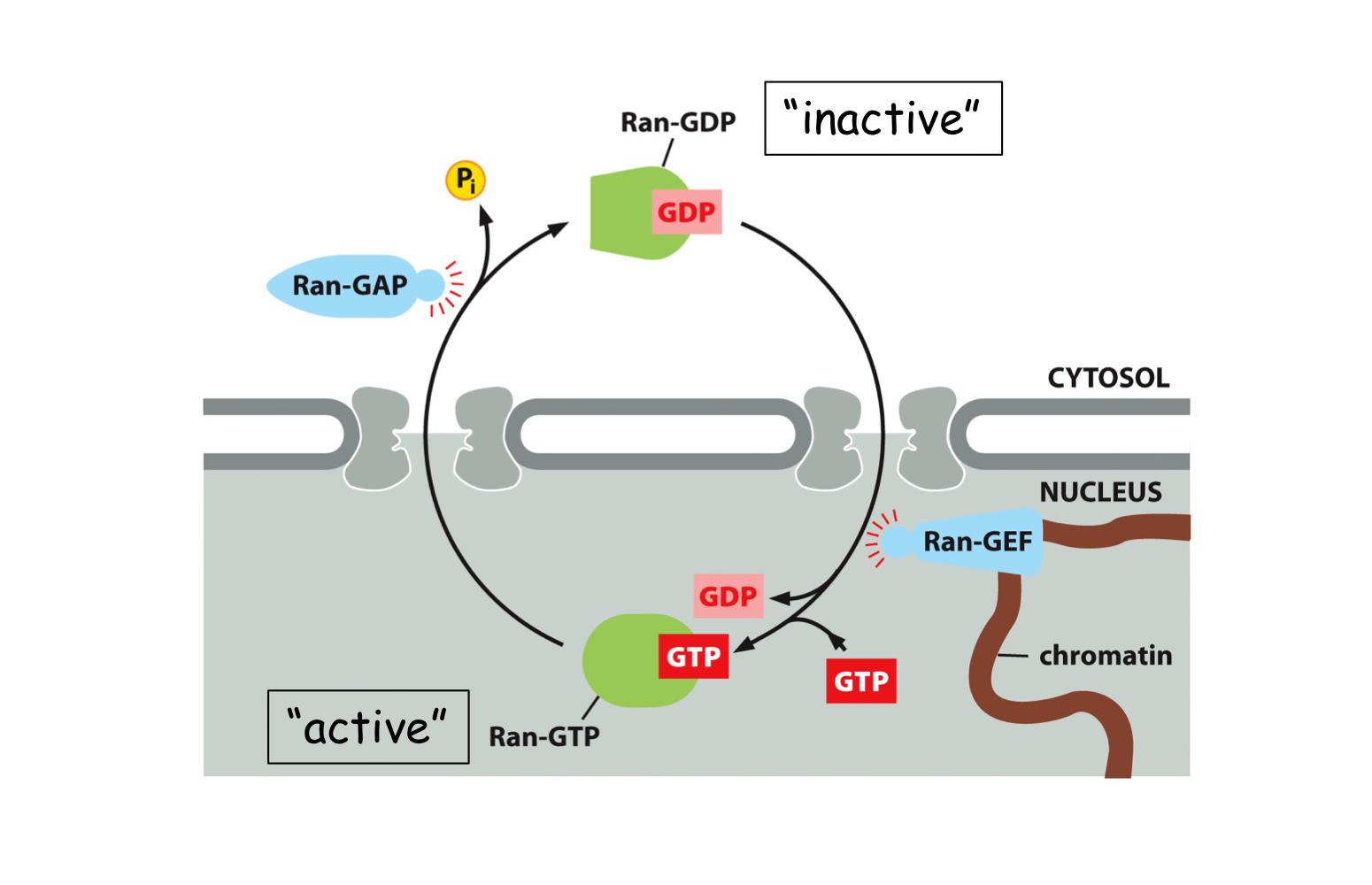
How is directionality ensured during nuclear transport?
The Ran “gradient”. The GTP-bound form only exists in the nucleus and the GDP-bound form only exists in the cytosol.
Mitochondrial import
N-terminal sorting sequence is required. If protein localizes to inter membrane spaces, a 2nd sorting sequence is needed
Mitochondrial import only occurs @ points where inner/outer membranes are in close contact
Matrix-targeting sequences
Matrix-targeting sequences are rich in hydrophobic, +charged & hydroxylated residues (Ser, Thr), but lack acidic residues. They tend to form amphipathic helices. Second sorting sequence used if the protein localizes to the intermembrane space
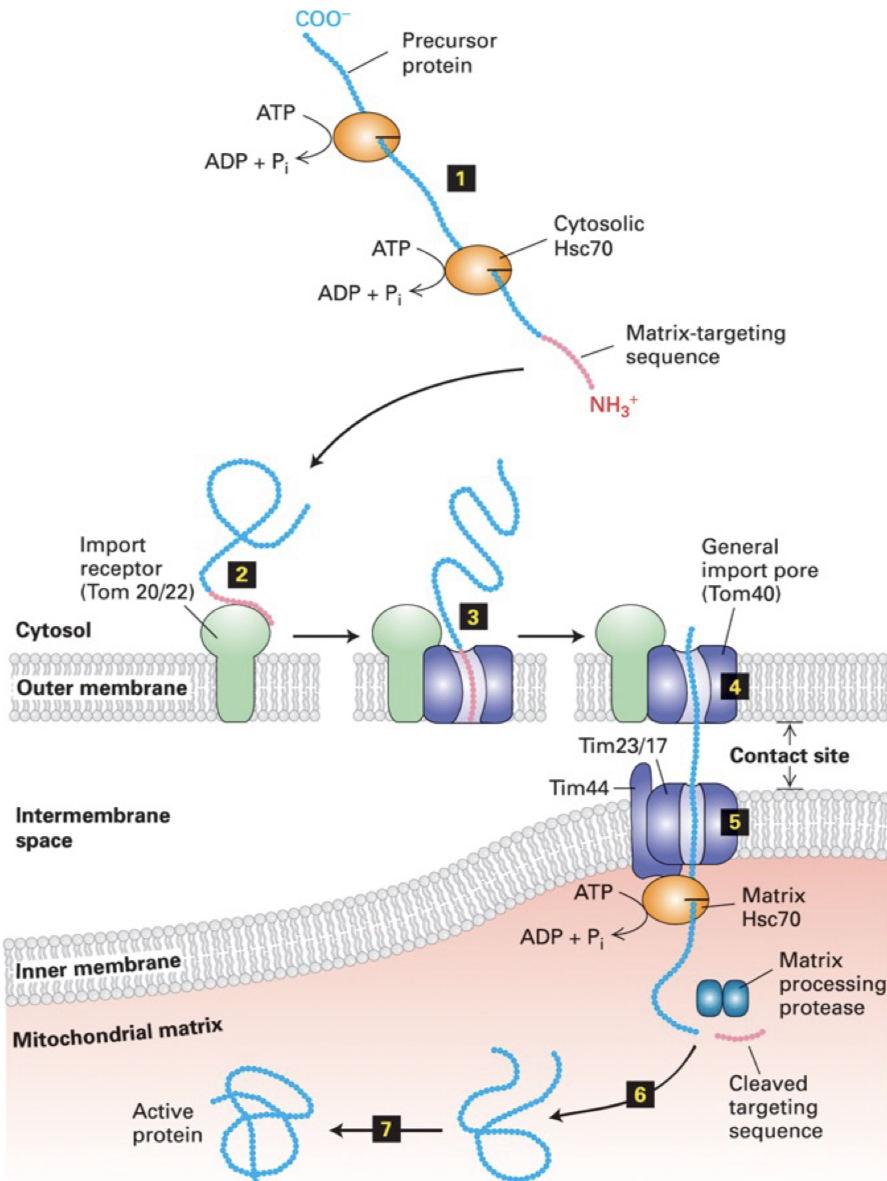
Post-translational import into mitochondrial matrix
precursor proteins are kept in unfolded state by cytosolic chaperone Hsc70 (requires energy in the form of ATP hydrolysis). Matrix-targeting sequence stays as an amphipathic a-helix
matrix-targeting sequence interacts w/ TOM20 or TOM22 receptor in the OM
receptor transfers protein to general import pore of OM composed of protein TOM40
@ outer/inner membrane contact points, protein passes thru import pore of IM composed of TIM23 & TIM17
Matrix Hsc70 binds to TIM44. ATP hydrolysis by this complex helps power translocation of protein → matrix
emerging matrix-targeting sequence is cleaved by a matrix protease
protein can now fold into its final shape, often aided by matrix chaperones
What is required for mitochondrial protein import?
The H⁺ electrochemical gradient generated by oxidative phosphorylation ensures that only actively respiring mitochondria can import. Hence, uncouplers block protein import
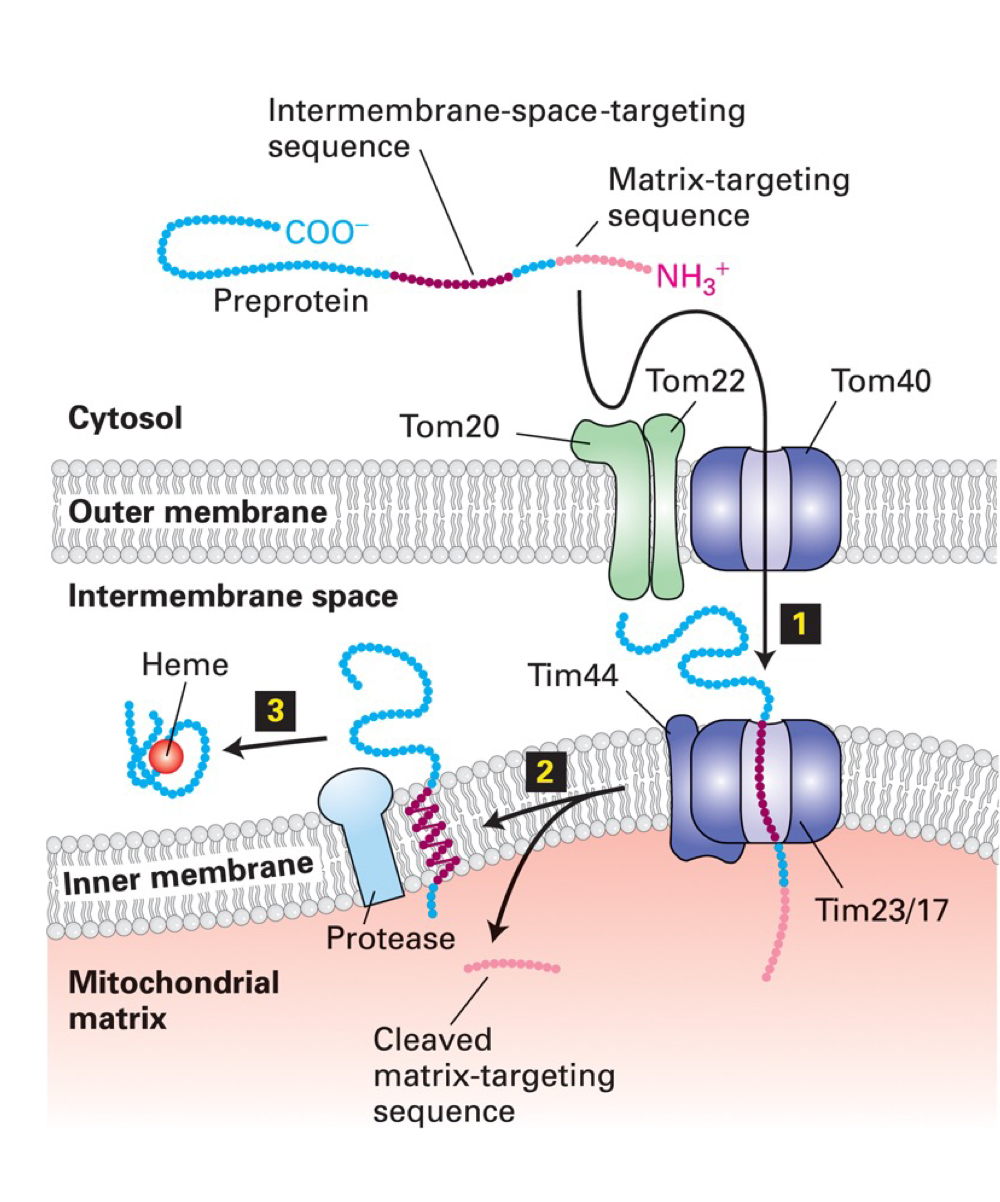
Mitochondrial import into intermembrane space
Targeting of proteins to intermembrane space requires second, hydrophobic targeting sequence that partially blocks protein from passing through TIM23/TIM17 import pore → stalled protein released from pore into membrane where membrane-anchored protease releases it into intermembrane space
(intermembrane sequence is hydrophobic enough to be inserted laterally into inner mito. membrane)
ER import
Most proteins that enter ER begin to be translocated across ER membrane before protein is completely synthesized (unlike proteins entering nucleus, mito. chloroplasts, peroxisomes). This requires that ribosomes synthesizing the protein are attached to ER (these ribosomes give rough ER its texture)
In the cytosol there are:
membrane-bound ribosomes that are attached to cytosolic surface of ER membrane and are synthesizing proteins translocated into ER
free ribosomes that are unattached to any membrane and are synthesizing all the other proteins
[membrane bound proteins of plasma membrane, Golgi, endsomes, lysosomes are inserted into ER membrane then transported using signals and AAs]
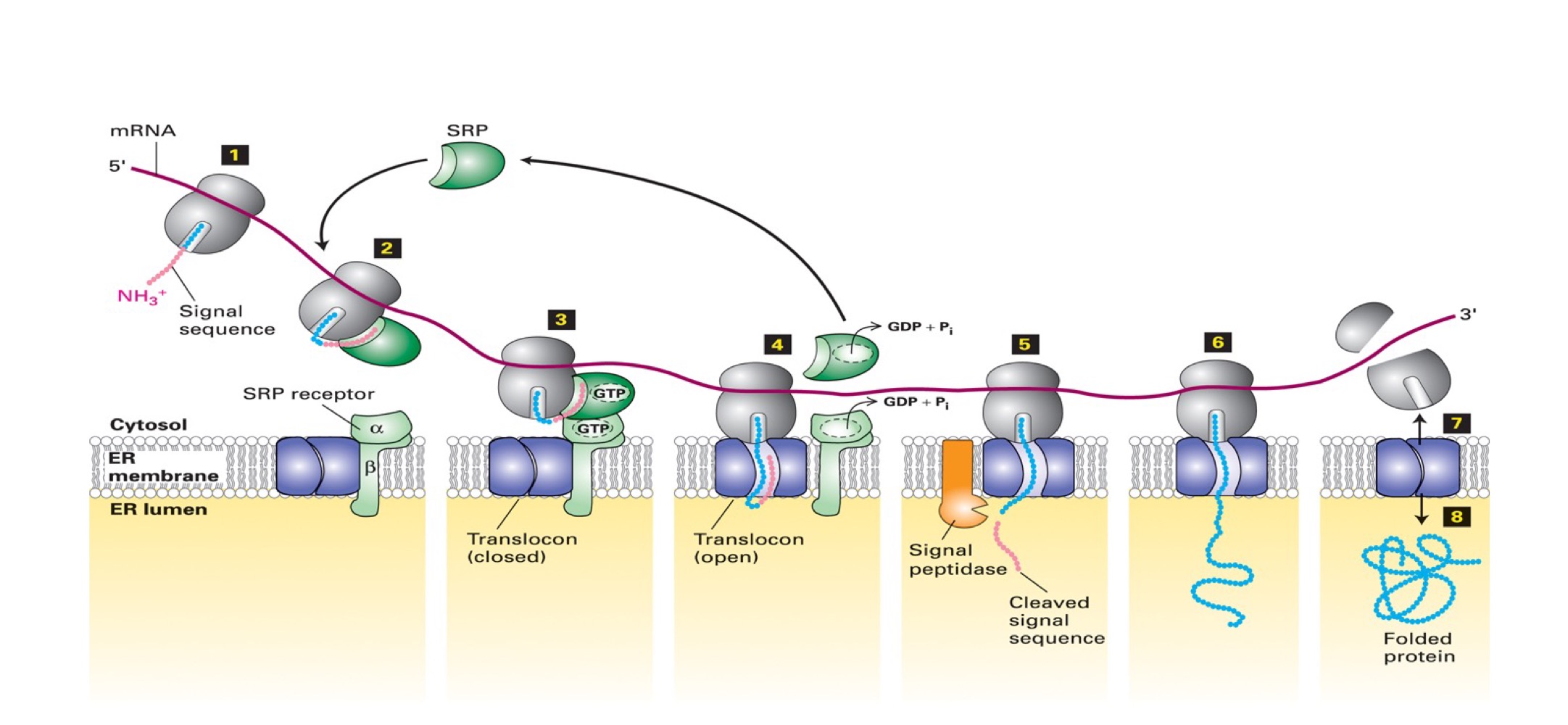
Steps for importing a soluble protein into ER
2. emerging polypeptide w/ its ER signal sequence is engaged by a complex of 6 proteins + RNA molecule SRP (signal recognition particle)
binding halts translation & delivers ribosome/polypeptide to ER *(abt 60 proteins need to be read before sequence is ejected)
SRP delivers ribosome/polypeptide to SRP receptor → interaction enhanced by binding of GTP to SRP & receptor
ribosome/polypeptide transferred to translocon, inducing it to open & receive polypeptide (enters as a loop). The hydrolysis of GTP by SRP & receptor free these factors for another importation
6. translation resumes → signal sequence cleaved by signal peptidase (membrane-bound protease). After digestion, rest of protein is synthesized and enters ER lumen
8. after the completed translation, ribosome is released causing translocon to close. New protein is folded
Membrane-anchored proteins
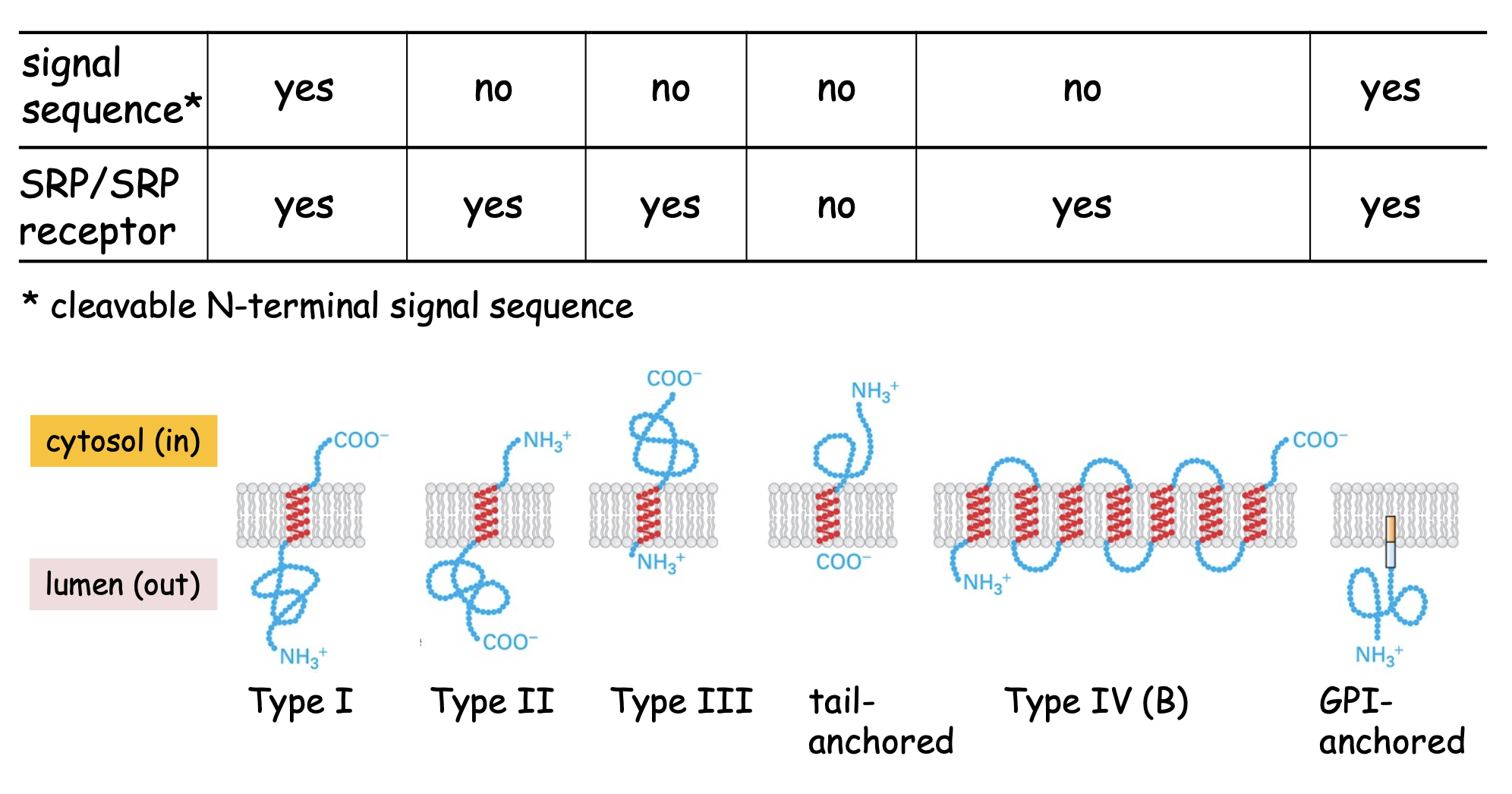
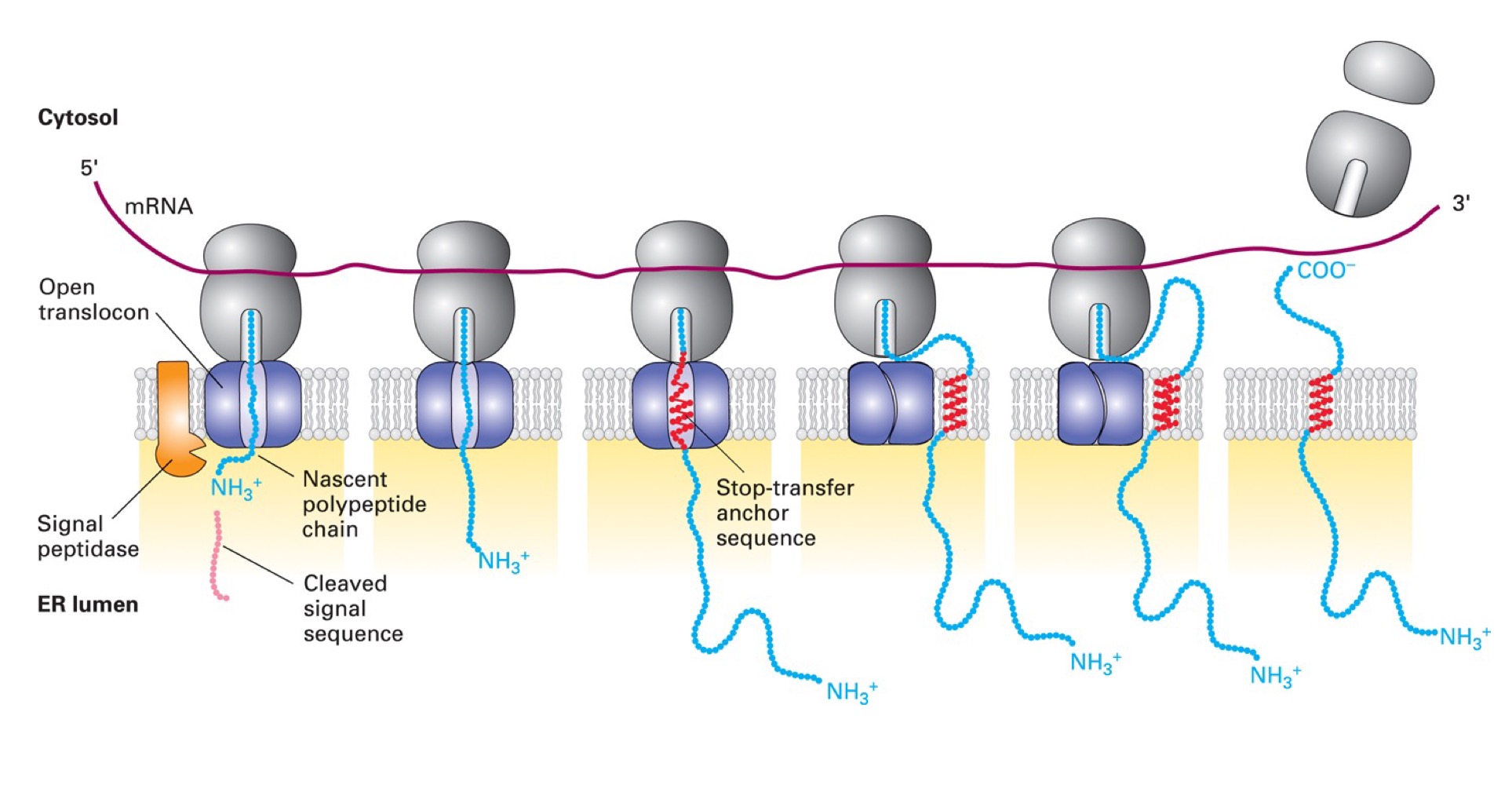
Type I membrane protein
Single pass, cleaved signal sequence @ N-terminus, uses SRP & receptor to get to ER membrane, Nₒ-Cᵢ
Proteins use a cleavable signal seq & STA (stop-transfer anchor) sequence that acts as membrane-spanning domain. Translocon opens to release hydrophobic stretch into membrane
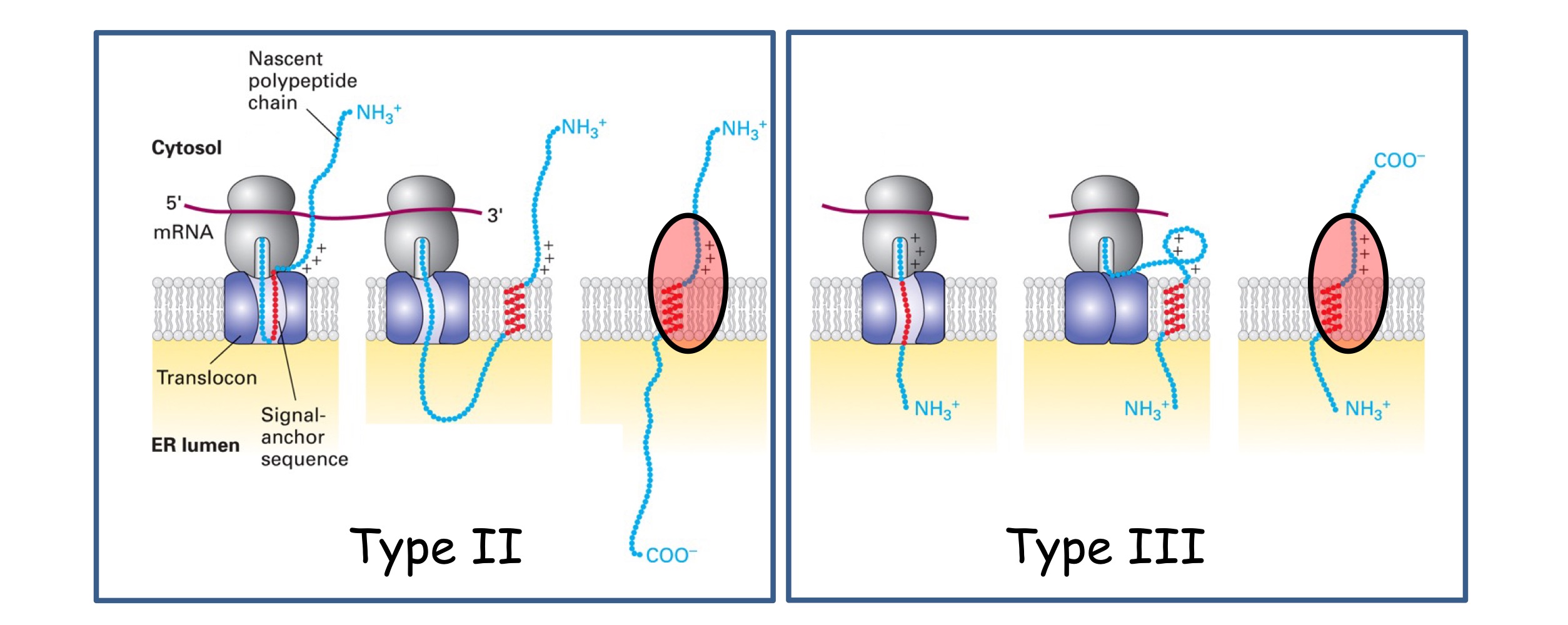
Type II & Type III
Single pass, no cleavable signal seq, hydrophobic membrane-spanning seq @ C-terminus, uses SRP & receptor to ER membrane, Nᵢ-Cₒ on type 2, Nₒ-Cᵢ on type 3
Orientation determined by +charged residues position relative to SA sequence :
between N-terminus & SA = TYPE II
between SA and C-terminus = TYPE III
These residues must remain in cytosol - this changes protein orientation
→ Use a SA (signal-anchor) sequence that acts as a dual signal sequence (directing protein to ER by SRP) and anchor OR membrane-spanning seq
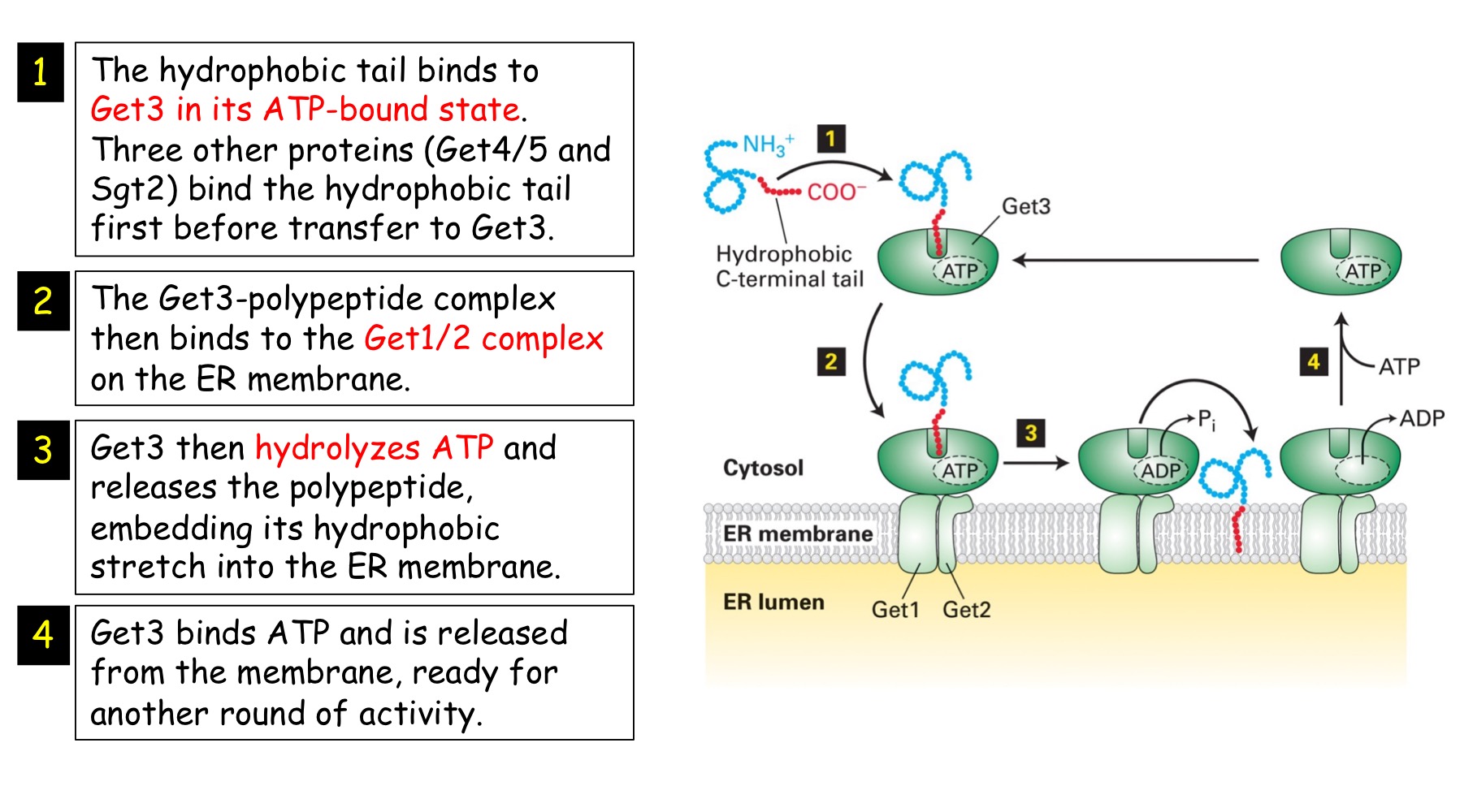
Tail-anchored
Single pass, no cleavable signal seq, hydrophobic membrane-spanning seq @ C-terminus, uses GET1/2/3 system to ER membrane, post-translational insertion, Nᵢ-Cₒ
→ they are inserted into ER after translation is complete since hydrophobic stretch that's inserted into bilayer needs to fully emerge from ribosome
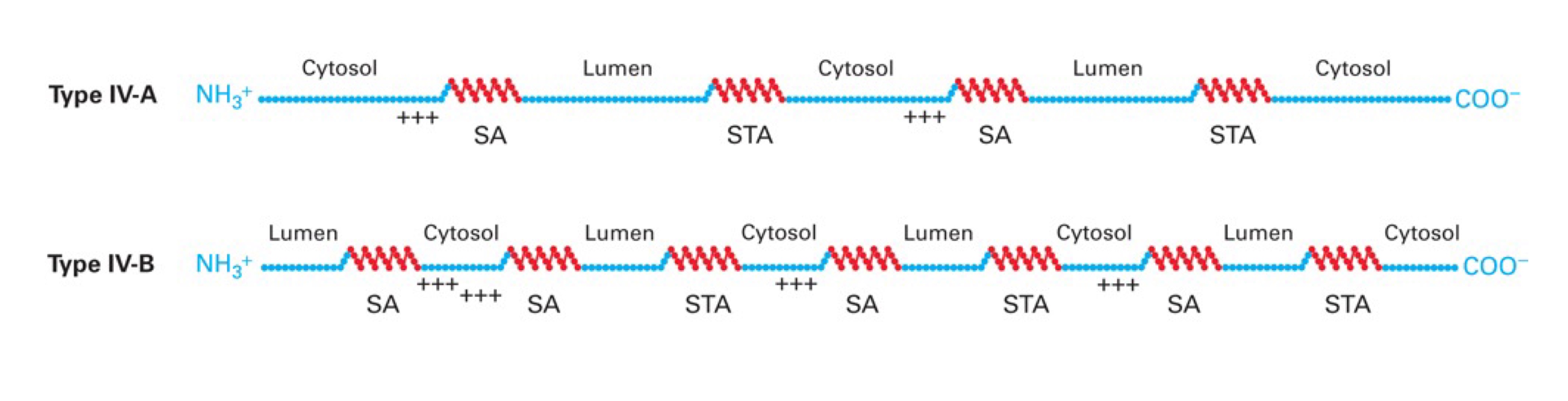
Type IV
Multi spanning, no cleavable signal seq, uses SRP & receptor for insertion of 1st membrane-spanning domain → IV-A are Nᵢ-Cᵢ ; IV-B are Nₒ-Cₒ
These proteins use combinations of Stop-Transfer Anchor sequences & Signal-Anchor sequences.
→ If the 1st SA sequence is a TYPE II (i.e. N-term+++SA) → protein will be Nᵢ like type II membrane proteins. If the 1st SA sequence is a TYPE III (i.e. SA+++C-term) → protein will be Nₒ like type III protein
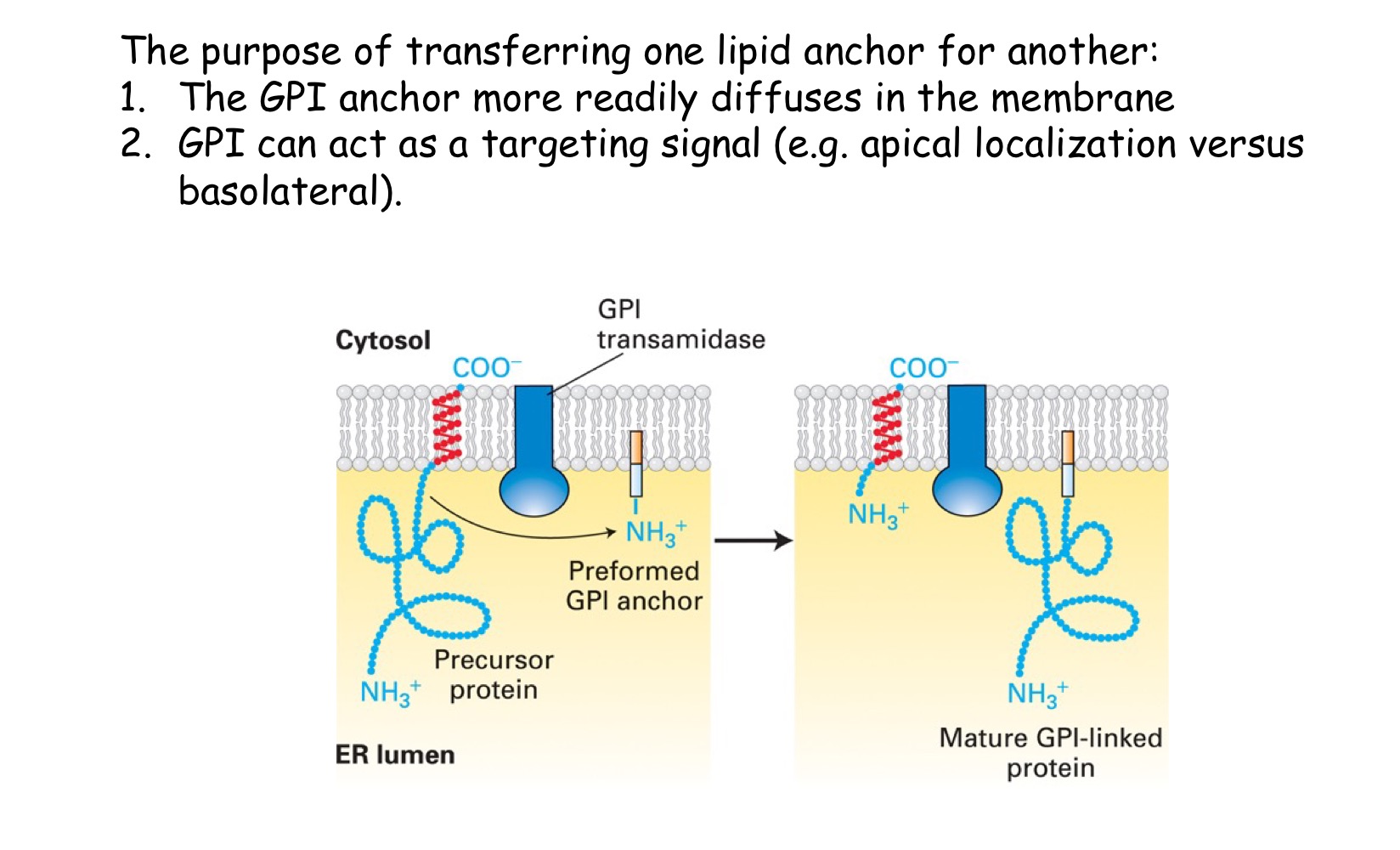
GPI-anchored
protein is lumenal, cleaved signal sequence @ N-terminus, uses SRP & receptor to get to ER membrane, anchored @ C-terminus to membrane → transferred to GPI anchor
These proteins insert into the ER like a Type I using a STA sequence. An enzyme (transamidase) then (i) cleaves the protein within the lumen of the ER and (ii) transfers it to the assembled GPI anchor (modified lipid - glycosylphosphatidylinositol)
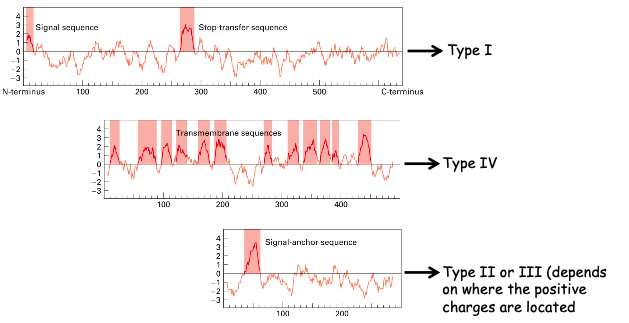
Hydropathic plots
Plots help determine the type of membrane protein. Hydropathic index for groups of 20 AAs are calculated and plotted against the protein sequence
the more hydrophobic the AA, the more positive the hydropathic index
★ we can’t further clarify the last plot since we don’t know where the positive charges are par rapport au SA sequence
☆ tail-anchored protein would only have one hydrophobic peak at the end of the plot
☆ GPI-anchored proteins enter similarly to TYPE I so a peak would be seen at both the very beginning and very end of the plot
ER is starting point for :
soluble proteins to be secreted from cell (e.g. hormones)
soluble proteins destined for lysosome, endosome, Golgi (e.g. acid hydrolase)
membrane proteins that will embed in Golgi, lysosome, endosome or plasma membrane (e.g. Na⁺/K⁺-ATPase)
Quality Control
Ensures that the proteins are properly modified, folded and assembled so they can be exported. Here are the main modifications:
Disulfide bond formation
Glycosylation
Folding
Proteolytic cleavage of amino-terminal signal sequences
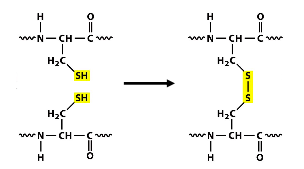
Disulfide bond formation
Covalent bond formation between thiol groups of cysteine residues on the same/different proteins (intra/inter molecular)
★ this bond formation is dependent on ER enzyme PDI (protein disulfide isomerase)
★ only modifies secreted proteins/ lumenal or EC domains of membrane proteins (cytosolic protein keratin is an exception → disulfide reduction in nair)
Bonds used to stabilize protein structure which is important for proteins that will be in extreme pH OR environments w/ high protease level (lysosome)
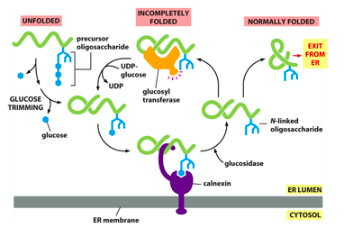
N-linked glycosylation
Sugar residues (oligosaccharides) are attached to nitrogen from asparagine residue [in consensus sequence Asn-X-Ser/Thr] by ER lumenal enzyme oligosaccharyl transferase
★ Precursor oligosaccharide is transferred to protein as cons. sequence emerges from translocon. The precursor is assembled step-by-step by lipid molecule dolichol (hydrophobic molecule w/ 75-95 C. it is phosphorylated at one end)
→ after glycosylation, 2 glucose residues removed, leaving behind 1 which is a Calnexin substrate. the core is not trimmed
![<p>Sugar residues (oligosaccharides) are attached to nitrogen from asparagine residue [in consensus sequence <strong><mark data-color="purple">Asn-X-Ser/Thr</mark></strong>] by ER lumenal enzyme <strong><span style="color: red">oligosaccharyl transferase</span></strong></p><p>★ Precursor oligosaccharide is transferred to protein as cons. sequence emerges from translocon. The precursor is assembled step-by-step by lipid molecule <strong><span style="color: purple">dolichol</span> </strong>(hydrophobic molecule w/ 75-95 C. it is phosphorylated at one end)</p><p>→<strong><span style="color: red"> </span></strong><em>after glycosylation, 2 glucose residues removed, leaving behind 1 which is a Calnexin substrate. the core is not trimmed</em></p>](https://knowt-user-attachments.s3.amazonaws.com/634111a2-46e0-4f51-bb5c-09679b75682f.jpeg)
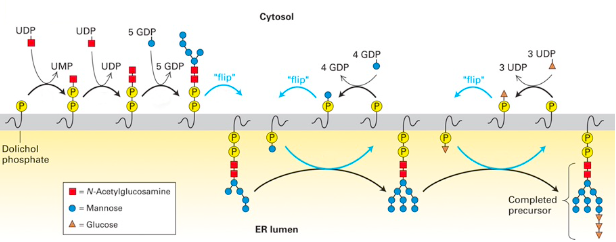
Glycosylation pathways
★ GlcNAc (🟥) is charged aka bound to UDP. An enzyme transfers 🟥 w/ a phosphate onto phosphorylated dolichol.
★ 2 GlcNAc residues & 5 mannose residues flip from cytosolic face to lumenal ER (using a transporter) since enzymes that continue to add sugars are there.
★ Mannose will be attached to dolichol in cytosol, which will flip inside to be added to precursor (x4). The same happens w/ 3 glucose molecules
☆ Sugar attachment to dolichol is mediated by nucleotides : UDP- GlcNAc, UDP-glucose, GDP-mannose
☆ Blocking N-linked glycosylation in ER can induce UPR (quality control will not deem protein properly folded if its not glycosylated) → tunicamycin blocks attachment of 1st GlcNAc residue to phosphorylated dolichol, which causes the rest of the process to stop
Roles of glycosylation
Promote folding of proteins (e.g. protein secretion is blocked for certain proteins when tunicamycin is used or if Asn/N is mutated)
Provide stability to proteins (e.g. some non-glycosylated proteins (fibronectin) are transported from the ER but are degraded faster)
Promote cell-cell adhesion on plasma membrane proteins (e.g. leukocyte-endothelial cell attachment during inflammatory response)
Act as a transport signal (e.g. mannose-6-phosphate directs proteins to the lysosome)
Protein folding
Molecular chaperones assist in folding by preventing aggregation of hydrophobic stretches of AAs.
Classical chaperones : Hsp70 (BiP), Hsp90, GRP94
Carbohydrate-binding chaperones : calnexin, calreticulin – bind to polypeptides that are monoglucosylated. Terminal glucose is removed and if folded the protein can exit the ER. If not, a glucosyltransferase adds one glucose back and the cycle repeats.
→ if mannose residues removed, protein is target for dislocation and degradation in cytosol by proteasome