Tumor Markers in solid tumor oncology
1/40
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
41 Terms
What can molecular testing due for cancer
Role of molecular testing in cancer:
Molecular testing can:
- Determine those patients who are at increased risk of cancer
- Determine tumor characteristics for:
o Diagnosis, prognosis, treatment
What are the 3 types of molecular marks (testing goals)
Types of molecular marks:
1. Diagnostic = establishes that a disease is present in the patient
- Presence of Philadelphia chromosome in CML
2. Prognostic = association with clinical outcomes such as survival or recurrence-free survival – independent of treatment
- Ex: p53 mutation indicates more aggressive cancer & HER/neu overexpression indicates growth and metastasis of breast cancer
3. Predictive = predict activity of a certain therapy response
- Ex: HER2/neu negative tumors do not respond to trastuzamab (Herceptin)
- Companion diagnostic: type of predictive marker that infers response to particular drug
o Positive test indicates drug may be used; test and drug are co-approved by FDA
o Ex: BRAF V600E positive melanoma is sensitive to vemurafenib and patient must be tested to be eligible for treatment
What are examples of solid tumors
Examples of solid tumors:
- Glioma, breast, colorectal, non-small cell lung cancer, Ewing sarcoma, Synovial sarcoma, & Rhabdomyosarcoma
How does signaling pathways work in cancer (basic answer)
Signaling pathways in cancer
- Signals from extracellular stimuli, like growth factors and hormones, bind to extracellular receptors and transmit through the cytoplasm and to the nucleus
- Results in cell proliferation or cell differentiation
o Typically highly controlled
- Proteins downstream from the cell surface receptor are turned on or off by protein modifications such a phosphorylation
o Enzymes that phosphorylate proteins are kinases
o Enzymes that take away or move a phosphate group are phosphatases and phosphorylases
What is germline testing
Germline testing detects inherited mutations present in all cells of the body and is used to identify cancer predisposition syndromes and assess inherited cancer risk in patients and family members (e.g., TP53 in Li–Fraumeni syndrome, BRCA1/BRCA2 in hereditary breast and ovarian cancer). Testing is performed on normal tissue such as blood.
What is somatic testing
Somatic testing detects acquired mutations present only in tumor cells and is used for diagnosis, prognosis, treatment selection, and monitoring of cancer. These mutations occur in oncogenes or tumor suppressor genes within the tumor and are not inherited (e.g., KRAS mutations affecting targeted therapy decisions).
Describe hereditary cancer and its testing
Hereditary cancer: RISK NOT DIAGNOSIS
- Important to remind the patient (and remember) that hereditary cancer mutations (germline) confer “risk” of cancer and are NOT a diagnosis of cancer
- Testing for hereditary cancer mutations is a genetic screening test to help in risk assessment
- Once level of risk is determined, more aggressive cancer screening can be done, such as more frequent mammograms, MRI, or ultrasound
o Occasionally patients with a very high risk may choose to undergo prophylactic surgeries (mastectomy or thyroid removal)
- Hereditary cancer testing using molecular methods is an opportunity to prevent disease, or at least catch it early
- A simple peripheral blood sample is typically all that is needed (EDTA purple-top tube)
Describe somatic cancer mutations and its testing
Somatic cancer mutations
- Sporadic cancer is caused by somatic mutations
- Somatic mutations typically ONLY OCCUR in the tumor
o Other cells in the body to not carry these mutations
o Mutations occur in tumor suppressor genes or oncogenes
- Somatic tumor mutation testing is done after the tumor is excised by surgery or occasionally on biopsy specimens
o Tumor is formalin fixed and paraffin imbedded
o Laboratory will slice into thin section and place onto slides for further analysis
o Tumor cells can be identified by a pathologist and then extracted by a molecular technologist for mutation testing
What are examples of hereditary cancer
Examples of hereditary cancer:
- BRCA1/2 mutations in breast cancer
- li-Fraumeni syndrome, p53 mutations
- Ataxia telangiectasia
- Von Hippel-Lindau
- Lynch syndrome
What is a screening
Screening = is done to for assessing cancer risk (likelihood of being diagnosed in the future)
- Disease is not yet present, molecular risk analysis provides actionable information for prevention or early detection
- Ex: BRCA testing is a screen, predicts risk of primary breast cancer or recurrence
How is a tumor diagnosed (test example)
Diagnosis of tumor
- Disease is present, molecular diagnosis establishes its characteristics to provide actionable information for treatment
- Ex: BRAF is a diagnostic test, which defines as an aggressive colon/lung tumor with potential for metastasis, as well as resistance to some chemotherapies
What does MDx (molecular diagnostics) of cancer tell us?
What does MDx of cancer tell us?
- The vast majority of molecular methods generally do not tell us “that the patient has cancer”; that is usually established by pathology, histology, immunohistochemistry prior to molecular testing. The MDx gives more details on cancer typing, prognosis, and prediction of response to therapy
Recall the steps of the cell cycle and relate to the function of tumor suppressors
The cell cycle consists of G1, S, G2, and M phases.
• G1 phase: cell growth
• S phase: DNA synthesis
• G2 phase: preparation for mitosis
• M phase: mitosis and cytokinesis
Tumor suppressor genes regulate cell cycle checkpoints at G1 → S and G2 → M transitions. They prevent inappropriate cell division by halting the cycle to allow DNA repair or by promoting apoptosis. Loss of tumor suppressor function allows uncontrolled progression through these checkpoints.
What is a oncogene and tumor suppressor gene mutation? Be able to predict in an example
How to predict mutations
- Oncogenes typically acquire gain-of-function mutations, resulting in constitutive activation of proteins that promote cell proliferation or survival (e.g., KRAS mutations causing constant signaling).
- Tumor suppressor genes typically undergo loss-of-function mutations, resulting in loss of cell cycle control, DNA repair, or apoptosis (e.g., TP53 inactivation).
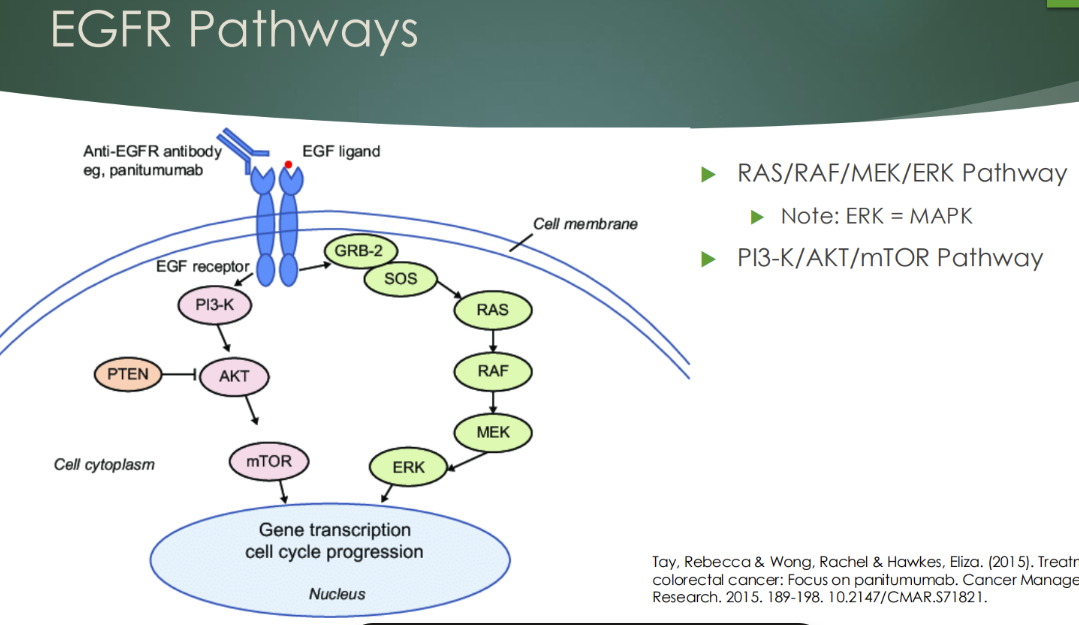
What is EGFR? What is KRAS? and its pathway simple
EGFR is a receptor tyrosine kinase that activates downstream signaling pathways, including the MAPK pathway.
KRAS acts downstream of EGFR as a GTP-binding protein, and BRAF functions further downstream in the same signaling cascade.
KRAS mutations are common oncogenic events and result in constitutive pathway activation independent of EGFR signaling. Mutations in KRAS and downstream genes such as BRAF influence response to targeted therapies, particularly tyrosine kinase inhibitors, and affect clinical treatment decisions. Tumor heterogeneity and pathway interactions complicate therapeutic response.
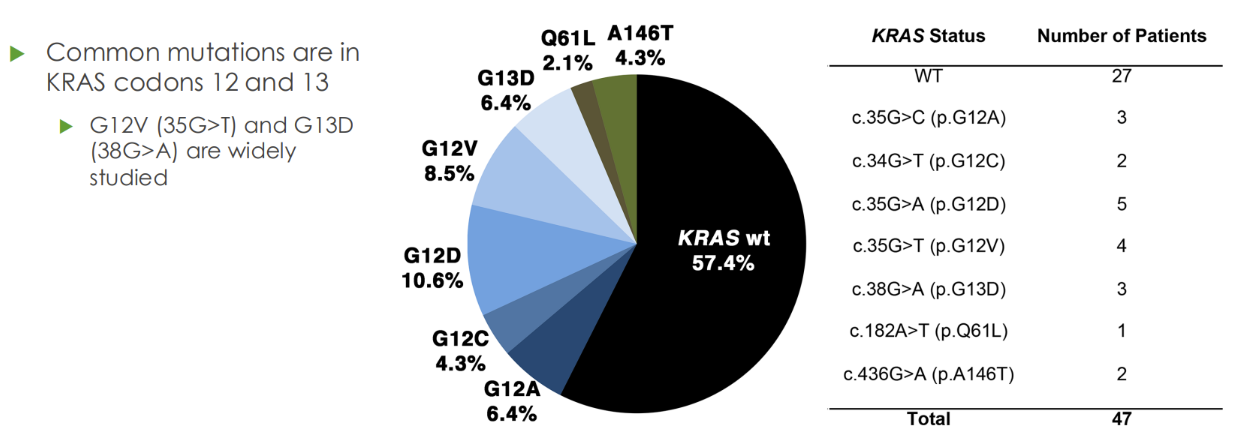
Describe KRAS in relation to cancer
KRAS (most common oncogene in all cancer)
- Part of RAS/RAF/MAPK pathway that is activated by stimulation of EGFR
- 40% of colon cancers are positive for KRAS mutations (won’t respond to sutuckanab)
- More than 95% of patients with KRAS mutations are unresponsive to EGFR – targeting monoclonal antibodies therapies, cetuximab and panitumumab
What are some recent treatments for KRAS G12C mutation
New treatments for KRAS G12C
- Until recently, there was no therapeutic target for mutated KRAS
- In 2021, the FDA approved the drug sotorasib (Lumakras), a RAS GTPase inhibitor, for patients with G12C mutations in NSCLC
o Particularly effective with anti-PD1 therapy (promotes cell death)
o Qiagen therascreen KRAS RGQ kit and Guardant260 CDx tests were approved as companioni diagnostics
Describe the mechanism of microsatellite instability, including the causative genes and expected laboratory results of MSI
Microsatellite instability
- Microsatellite instability results from defects in the DNA mismatch repair (MMR) system, which normally corrects replication errors such as base mismatches and insertion/deletion loops.
- Causative genes include MLH1, MSH2, MSH6, and hPMS2. Dysfunction of these genes leads to accumulation of replication errors in microsatellite regions, producing new alleles over successive cell divisions.
- Laboratory results show increased numbers of alleles in tumor DNA compared with normal DNA when microsatellite loci are analyzed by PCR and electrophoresis. Tumors are classified as MSI-H, MSI-L, or MSS based on the proportion of unstable loci.
Describe EGFR, KRAS, and BRAF mutations in colorectal cancer
EGFR RAS/RAF/MAPK pathway in colorectal cancer
- Tumors of the large intestine
- EGFR is overexpressed/amplified (extra copies on cell surface) in colon cancer
o Treated with EGFR monoclonal antibodies (cetuximab and related drugs)
- KRAS mutation
o Indicates that EGFR drugs will not work
- In KRAS wildtype, a BRAF mutation
o Indicates that EGFR drugs will not work
- KRAS and BRAF mutations turn on the pathway even when EGFR is blocked
Describe BRAF in colorectal cancer
BRAF (BRAF, V600E) in colorectal cancer
- Mutation makes RAF constitutively active, circumventing EGFR-directed therapy effectiveness
- BRAF testing recommended if KRAS is found to be wildtype
- Mutations in KRAS and BRAF are mutually exclusive – almost never coexist in the same tumor
- Although colorectal cancers with the BRAF V600E mutations won’t respond to cetuximab, they may respond to the melanoma drug vemurafenib (companion diagnostic for melanoma; some evidence of response in colorectal cancer)
Describe EGFR and KRAS mutations in NSCLC?
EGFR/RAS/RAF pathway in non-small cell lung cancer (NSCLC)
- NSCLC are lung cancers that are not “small cell”
o ~85% of lung cancers are NSCLC
o Small cell lung cancers are more aggressive, but also more responsive (but also cancer often returns)
- NSCLC may have EGFR mutations
o Cause constitutively active tyrosine kinase activity
o Tumors with such mutations show high sensitivity to EGFR tyrosine kinase inhibitors gefitinib and erlotinib
Most common mutations: LREA exon 19/20 deletion and exon 21 point mutation L858R (these are ACTIVATING mutations)
Wildtype EGFR tumors do NOT respond to EGFR inhibitors
- KRAS in NSCLC
o >20% of patients
o Mutually exclusive from EGFR mutations
Mutant KRAS means wildtype EGFR
Also means wildtype BRAF
o Confers resistance to gefitinib and erlotinib TKIs
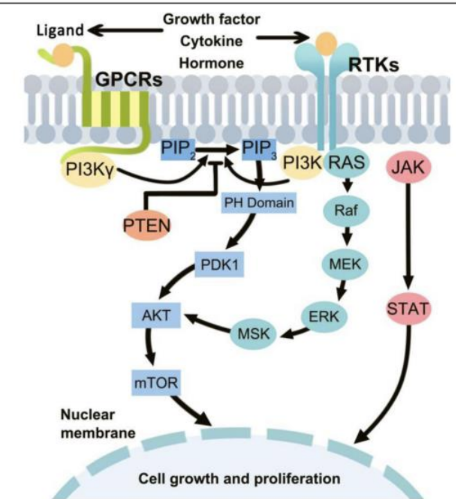
Describe the PI3K/AKT/mTOR pathway in many cancers
PI3K/AKT/mTOR pathway in many cancers
- PI3K signaling is deregulated in a wide variety of cancers
o Breast, gastric, ovarian, colorectal, prostate, glioblastoma, endometrial
- Alterations include:
o Loss or inactivation of tumor suppressor PTEN
o Mutations or amplification of PI3K (PIK3CA gene)
o Activation of tyrosine kinase growth factor receptors upstream of PI3K
- Drugs include a variety of FDA-approved and experimental types
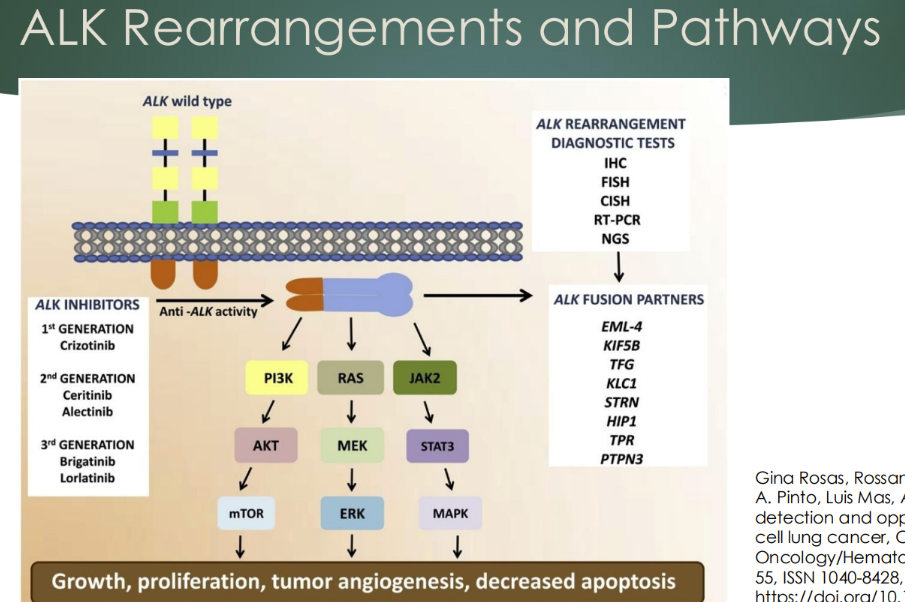
Describe an ALK mutation in NSCLC
ALK in NSCLC
Alk gene fusion
- Chromosomal mutation creating a fusion protein with the EML4 gene (or others)
o ALK can activate signaling pathways, including MAPK and PI3K
- The first drug crizotinib (a tyrosine kinase inhibitor) was approved with a companion diagnostic for the translocation (FISH)
o Newer generation drugs: ceritinib, alectinib, ensartinib, lorlatinib
- Detected by FISH, CISH, or NGS
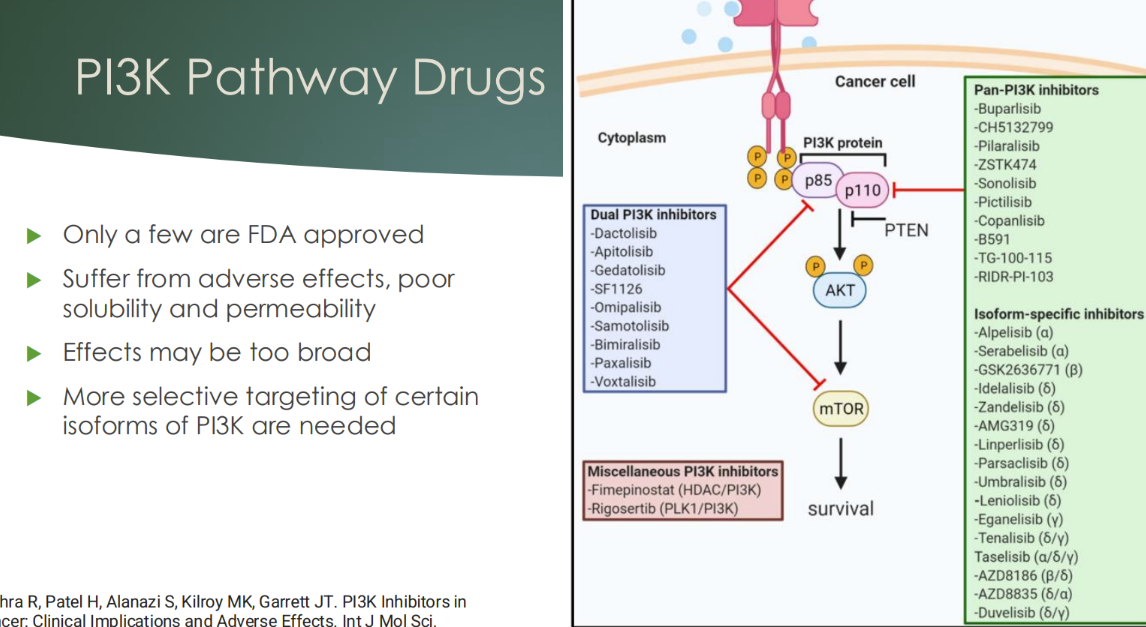
Describe the situation with PI3K pathway and drugs
only a few FDA approved
suffer from adverse effects, poor solubility and permeability
effects may be too broad
more selective targeting of certain isoforms of PI3K are needed
What are the molecular targets of HER2/neu, EGFR, KRAS, EWS-FLI1, SYT-SSX, PAX3, TP53, RET, and KIT mutations
Examples of molecular targets described in the PDF include:
• HER2/neu amplification in breast cancer (IHC, FISH)
• EGFR mutations in lung cancer
• KRAS mutations in lung, colon, thyroid, and skin cancers
• EWS-FLI1 translocation in Ewing sarcoma
• SYT-SSX translocation in synovial sarcoma
• PAX3-FKHR / PAX7-FKHR translocations in rhabdomyosarcoma
• TP53 mutations across many tumor types
• RET and ROS1 rearrangements in lung and thyroid cancers
• KIT mutations in gastrointestinal stromal tumors
Discuss the role of p53 mutations across multiple cancer classifications
TP53 mutations occur in approximately 50% of all cancers and are found across many cancer types, including breast, lung, colon, and leukemias. The p53 protein normally halts the cell cycle in response to DNA damage, allowing repair before replication. Loss of p53 function permits replication of damaged DNA, leading to genomic instability. Mutant p53 protein accumulates in cells and is associated with poor prognosis.
KRAS mutation treatment options
- More than 95% of patients with KRAS mutations are unresponsive to EGFR – targeting monoclonal antibodies therapies, cetuximab and panitumumab
- In 2021, the FDA approved the drug sotorasib (Lumakras), a RAS GTPase inhibitor, for patients with G12C mutations in NSCLC
o Particularly effective with anti-PD1 therapy (promotes cell death)
o Qiagen therascreen KRAS RGQ kit and Guardant260 CDx tests were approved as companioni diagnostics
- KRAS mutation
o Indicates that EGFR drugs will not work
- KRAS and BRAF mutations turn on the pathway even when EGFR is blocked
- Mutations in KRAS and BRAF are mutually exclusive – almost never coexist in the same tumor
- KRAS in NSCLC
o >20% of patients
o Mutually exclusive from EGFR mutations
Mutant KRAS means wildtype EGFR
Also means wildtype BRAF
o Confers resistance to gefitinib and erlotinib TKIs
EGFR mutation treatment options
- EGFR is overexpressed/amplified (extra copies on cell surface) in colon cancer
o Treated with EGFR monoclonal antibodies (cetuximab and related drugs)
- NSCLC may have EGFR mutations
o Cause constitutively active tyrosine kinase activity
o Tumors with such mutations show high sensitivity to EGFR tyrosine kinase inhibitors gefitinib and erlotinib
Most common mutations: LREA exon 19/20 deletion and exon 21 point mutation L858R (these are ACTIVATING mutations)
Wildtype EGFR tumors do NOT respond to EGFR inhibitors
BRAF mutation treatment options
- In KRAS wildtype, a BRAF mutation
o Indicates that EGFR drugs will not work
- KRAS and BRAF mutations turn on the pathway even when EGFR is blocked
- Mutations in KRAS and BRAF are mutually exclusive – almost never coexist in the same tumor
- Although colorectal cancers with the BRAF V600E mutations won’t respond to cetuximab, they may respond to the melanoma drug vemurafenib (companion diagnostic for melanoma; some evidence of response in colorectal cancer)
What are some treatment options of ALK gene fusion mutations
- Chromosomal mutation creating a fusion protein with the EML4 gene (or others)
o ALK can activate signaling pathways, including MAPK and PI3K
- The first drug crizotinib (a tyrosine kinase inhibitor) was approved with a companion diagnostic for the translocation (FISH)
o Newer generation drugs: ceritinib, alectinib, ensartinib, lorlatinib
Associate companion diagnostics (key 1st generation drugs) with gene target and mutation
Gene Target | Specific Mutation / Alteration | Cancer Type | 1st Generation Drug | Companion Diagnostic Association |
BRAF | V600E mutation | Melanoma (also colorectal cancer) | Vemurafenib | Patients must test positive for BRAF V600E to be eligible for therapy |
EGFR | Exon 19 deletions; L858R point mutation | Non-small cell lung cancer (NSCLC) | Gefitinib, Erlotinib | EGFR mutation testing required; wildtype EGFR tumors do not respond |
ALK | ALK gene rearrangement (e.g., EML4-ALK fusion) | NSCLC | Crizotinib | ALK translocation must be detected (FISH or equivalent) prior to treatment |
KRAS | G12C mutation | NSCLC | Sotorasib (Lumakras) | FDA-approved companion diagnostics (Qiagen therascreen KRAS RGQ, Guardant360 CDx) identify eligible patients |
-ab = monoclonal drug
-ib = Tyrosine Kinase Inhibitors (EGFR) drug
Made by chat lol
What are the steps of FFPE DNA extraction with Qiagen
FFPE DNA extraction (Qiagen)
1. Scrape tissue section using clean scalpel blade
2. Remove paraffin: Dissolve in xylene and removed
3. Lyse: lysed under denaturing conditions with proteinase K
4. Heat: Incubation at 90 degrees C reverses formalin crosslinking
5. Bind: DNA binds to membrane and contaminants flow through
6. Wash: residual contaminants washed away
7. Elute: pure, concentrated DNA elute from membrane
What are the steps of FFPE DNA extration with Ion AmpliSeq
FFPE DNA (Ion AmpliSeq)
1. Using a 20uL pipette tip, add 10uL of transfer solution onto region of interest of the slide-mounted FFPE tissue section
2. Spread transfer solution, then scrape and break up the tissue with pipette tip. The tissue is a slurry of fine particles
3. Pipet the slurry from the slide into a 0.2ml tube
4. Add direct reagent and mix. Incubate 65 degrees for 15 min then 95 degrees for 10 min
5. Centrifuge briefly. The bottom phase contains extracted DNA and can be used for sequencing

Describe the Idylla platform for FFPE or plasma
Idylla platform for FFPE or plasma
- Can directly add FFPE scrape or plasma sample into a cartridge
- Idylla instrument does sample extraction and real-time PCR all in one cartridge
- Assays available for...
o ALK/ROS/RET, NTRK gene fusions
o EGFR mutations
o MSI loci
o KRAS and BRAF

Define exosomes in context of liquid biopsy
Exosomes are cell-derived vesicles released into circulation that carry nucleic acids and other molecules. In liquid biopsy, exosomes serve as a source of tumor-derived genetic material that can be analyzed for mutations, gene expression, and other molecular characteristics without requiring invasive tissue biopsy.
How deos Qiagen therascreen RGQ detect KRAS mutations
Qiagen therascreen KRAS RGQ
- Real-time PCR kit using RotorGene Q
- Based on amplification refractory mutation system (ARMS) PCR
o 7 mutation-specific reactions in codons 12 and 13
o 1 control reaction in exon 4 (wildtype)
- Uses scorpion probes for detection
- Can detect as little as about 5% of mutations in a mixture of wildtype
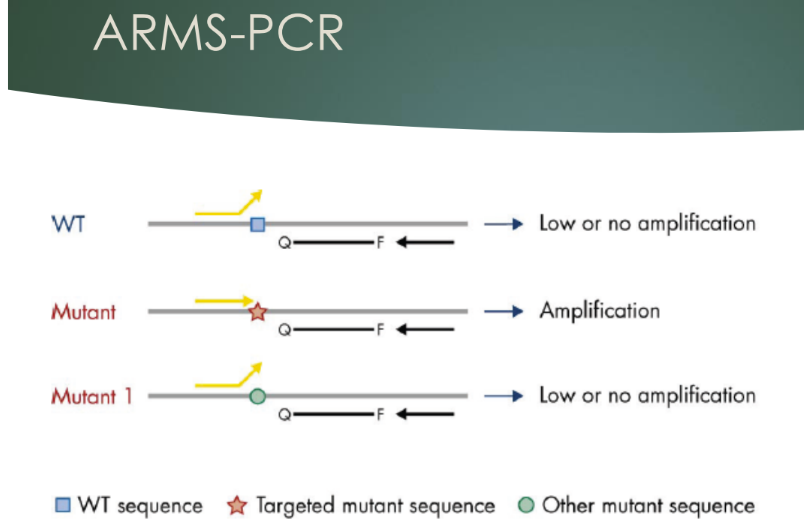
What does ARMS-PCR do
- Mismatches in primer makes them allele-specific
- In the therascreen assay, the ARMS-PCR is used to preferentially amplify the mutated targets, to increase the signal to noise ratio against the wildtype (noise)
- In this case the perfectly matched mutation primers amplify with high efficiency while the wildtype primers show only low-level background
What is the Guardant 360CDx
Guardant 360CDx
- NGS based diagnostic for multiple targets found in circulating cell-free DNA from plasma of peripheral blood
- To be used in patients already diagnosed with cancer (solid tumor)
- FDA-approved companion diagnostic for NSCLC:
o EGFR exon 19 deletions; and mutations L858R and T790M (osimertinib)
o EGFR exon 20 insertions (aminvantamab)
o KRAS G12C (sotorasib)
What is the workflow of the Guardant 360CDx
Guardant 360CDx workflow
1. Whole blood collection and shipping
- 5 ml blood
2. Plasma isolation and cfDNA extraction
- Centrifuge to isolate plasma
- cfDNA extracted
3. Library preparation and enrichment
- 5 to 30 ng for library, enriched by hybridication capture
4. DNA sequencing
- Illumina NextSeq 550 platform
5. Data analysis and reporting
- Custom bioinformatics pipeline
- Results are detected or not detected (qualitative)
What are specialized extraction methods
Specialized extraction methods:
- Cell-free DNA extraction from blood/plasma
- Formalin-fixed paraffin-embedded tissue sections
What is the process of cell-free DNA extraction
Cell-free DNA extraction
1. To separate plasma, centrifuge whole blood at 300-1600 x g for 10-20 min at room temp
2. Remove upper plasma layer and transfer to new conical tube
3. Centrifuge the plasma at 5000-16000 x g for 10 min
4. Extract using cfDNA kit or instrument
- Qiagen QIA amp or QIA symphony kits
- MagMax kit on KingFisher instrument