Midterm 1 Biochemistry
1/107
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
108 Terms
What are irreversible inhibitors and how do they work?
Irreversible inhibitors permanently inactivate enzymes by reacting with them, often as toxins (e.g., sarin gas causes paralysis by inhibiting acetylcholinesterase).
What are reversible inhibitors and what types exist?
Reversible inhibitors bind and dissociate easily; they can be competitive, uncompetitive, or noncompetitive, often mimicking the substrate or product.
How do reversible inhibitors block enzyme activity?
They bind to either the free enzyme (blocking substrate binding) or to the enzyme-substrate complex (blocking the reaction).
What happens in competitive inhibition?
A competitive inhibitor binds the active site of the enzyme, blocking the substrate and reducing native substrate binding.
Simple Example: It's like two people trying to sit in the same chair — only one can occupy it at a time!
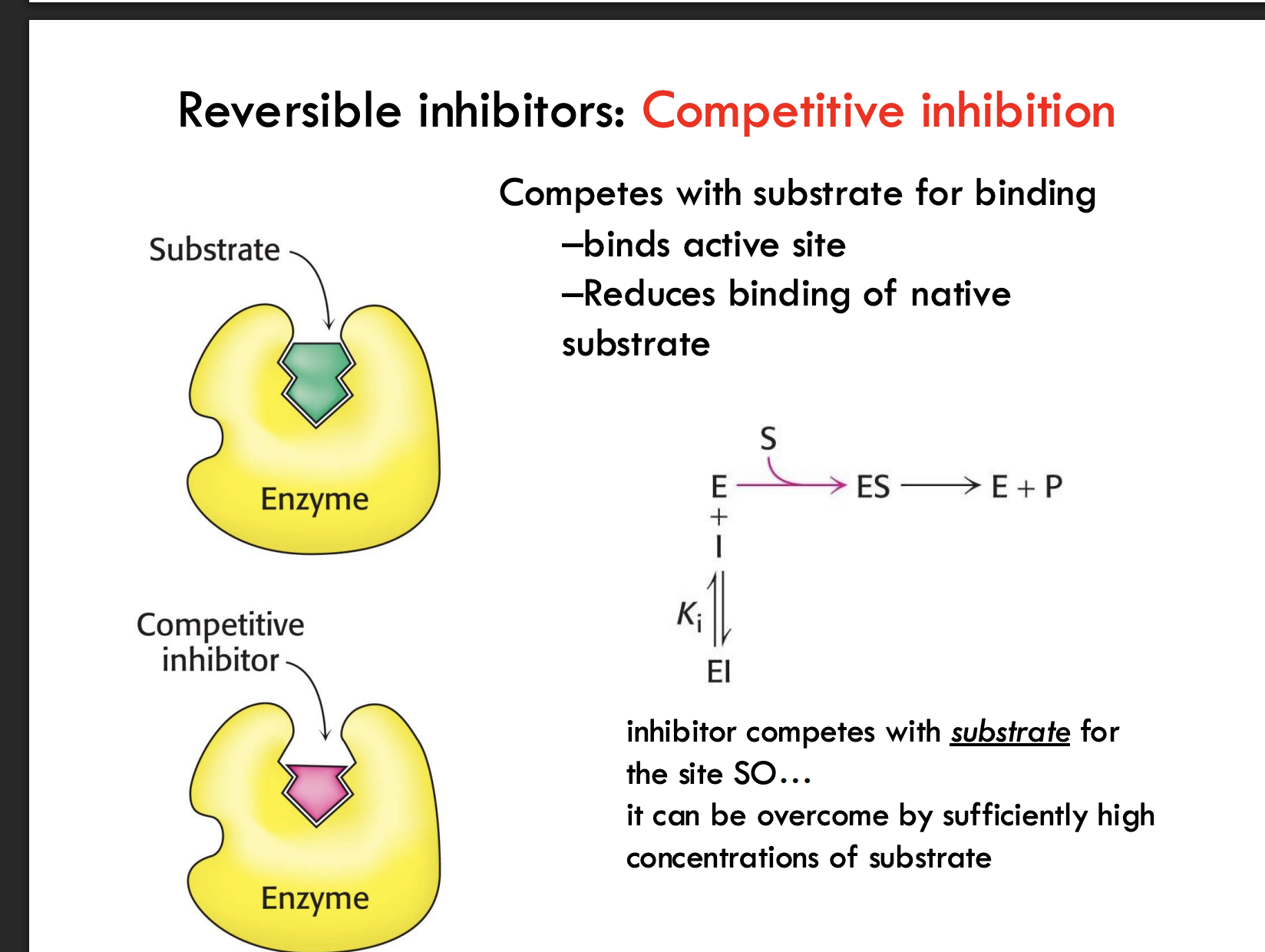
How can competitive inhibition be overcome?
By adding a high concentration of substrate, outcompeting the inhibitor for binding to the active site.
Simple Example: If enough people rush for the chair (substrate), the competitor (inhibitor) gets pushed aside.
How does competitive inhibition affect Vmax?
Vmax stays unchanged because enough substrate can still outcompete the inhibitor.
Simple Example: Imagine a race where no matter how many hurdles you put, the fastest runner (substrate) will still eventually win if you let enough of them race.
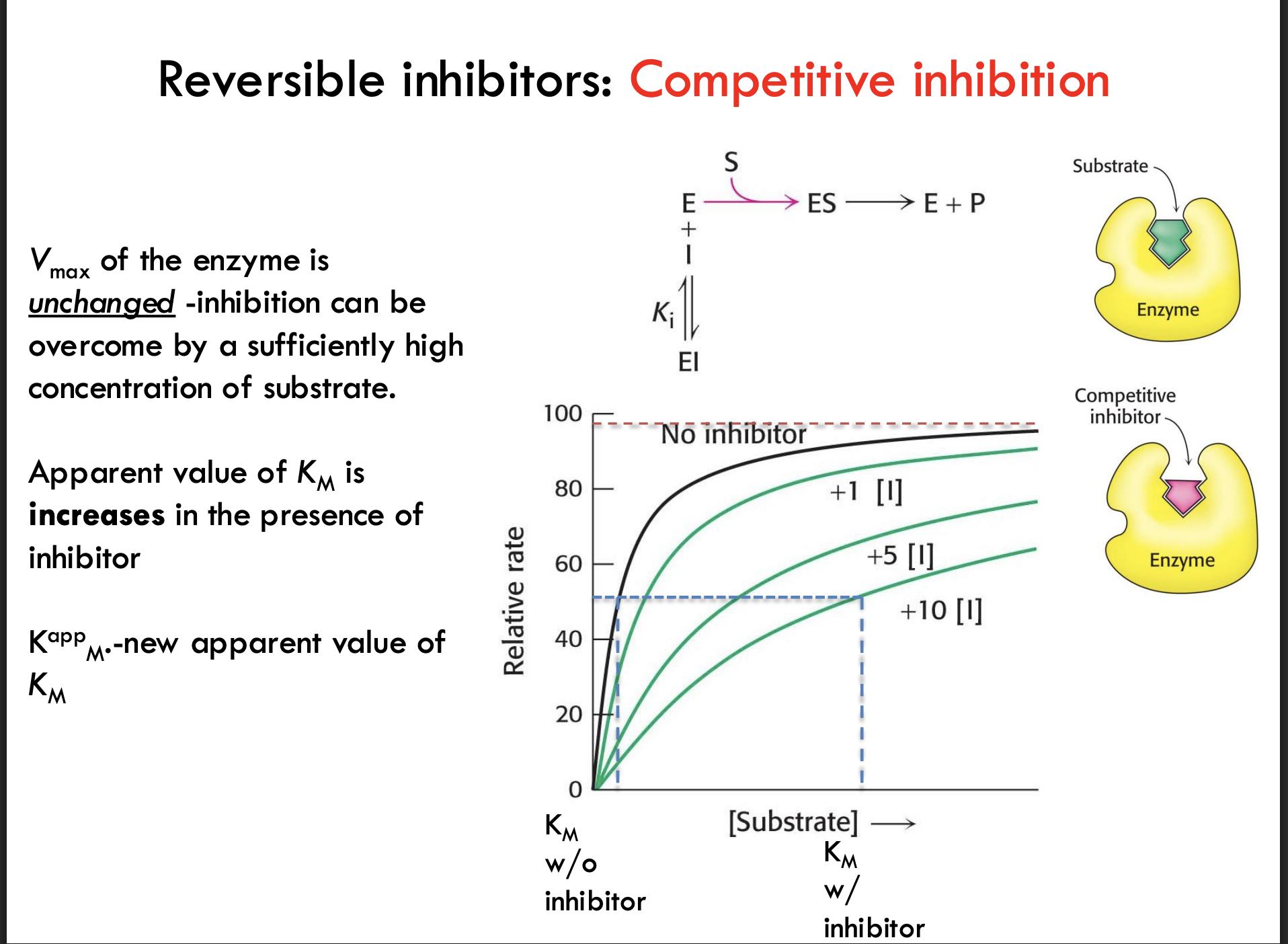
How does competitive inhibition affect Km?
The apparent Km increases, meaning it looks like it takes more substrate to reach 1/2 Vmax.
Simple Example: It’s like needing more people to push a heavy door when someone is leaning against it (the inhibitor).
What does "Kₘᵃᵖᵖ" represent in competitive inhibition?
Kₘᵃᵖᵖ is the new, higher Km value measured when a competitive inhibitor is present.
Simple Example: Without the blocker, you need 5 people to push the door open (Km); with a blocker (inhibitor), you now need 10 people (Kmᵃᵖᵖ).
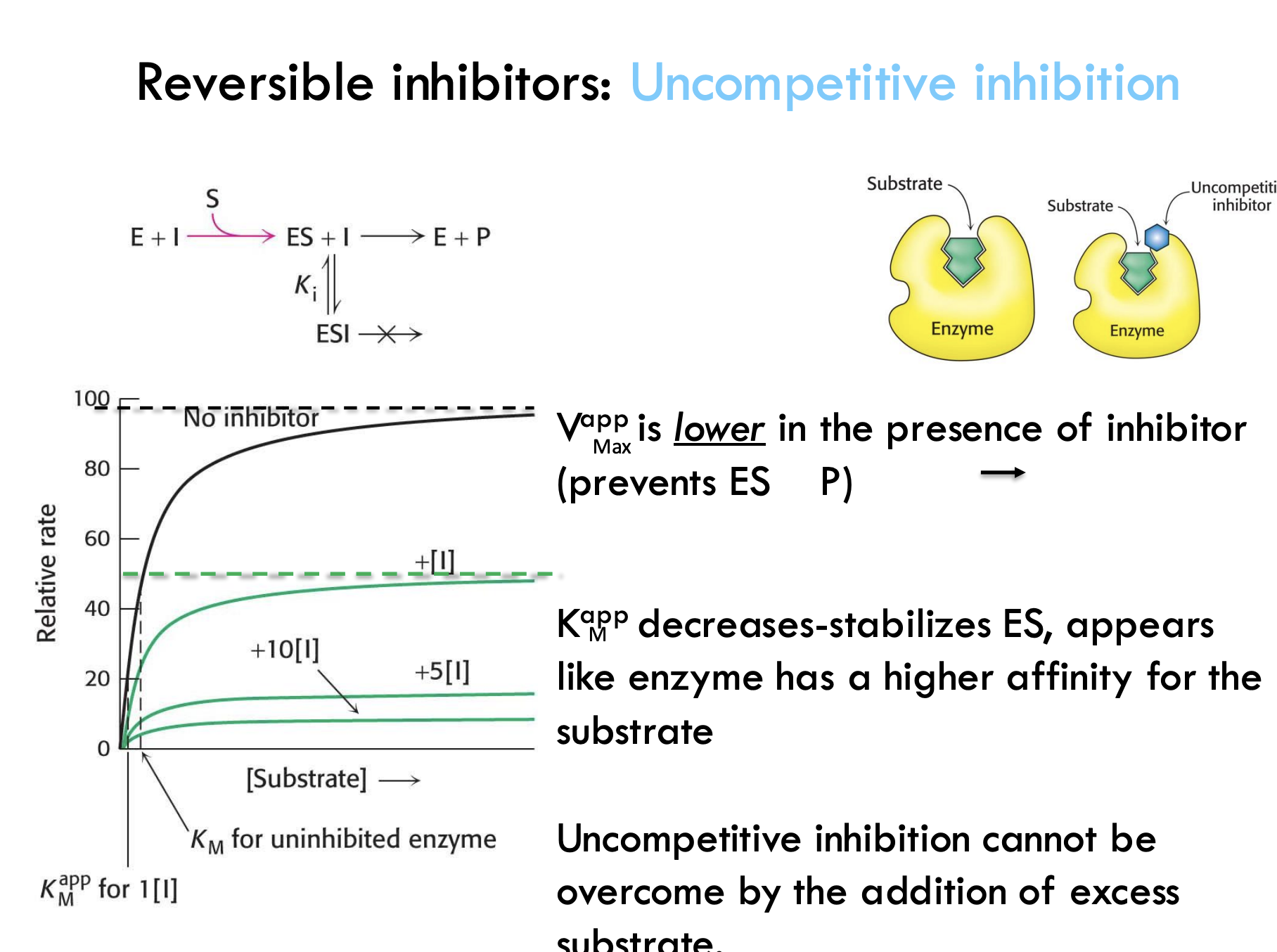
What happens during uncompetitive inhibition?
The inhibitor binds only after the substrate is already bound and locks the enzyme-substrate complex, stopping the reaction.
Simple Example: Imagine someone putting a lock on a vending machine after you already picked your snack — you can't get it out anymore.
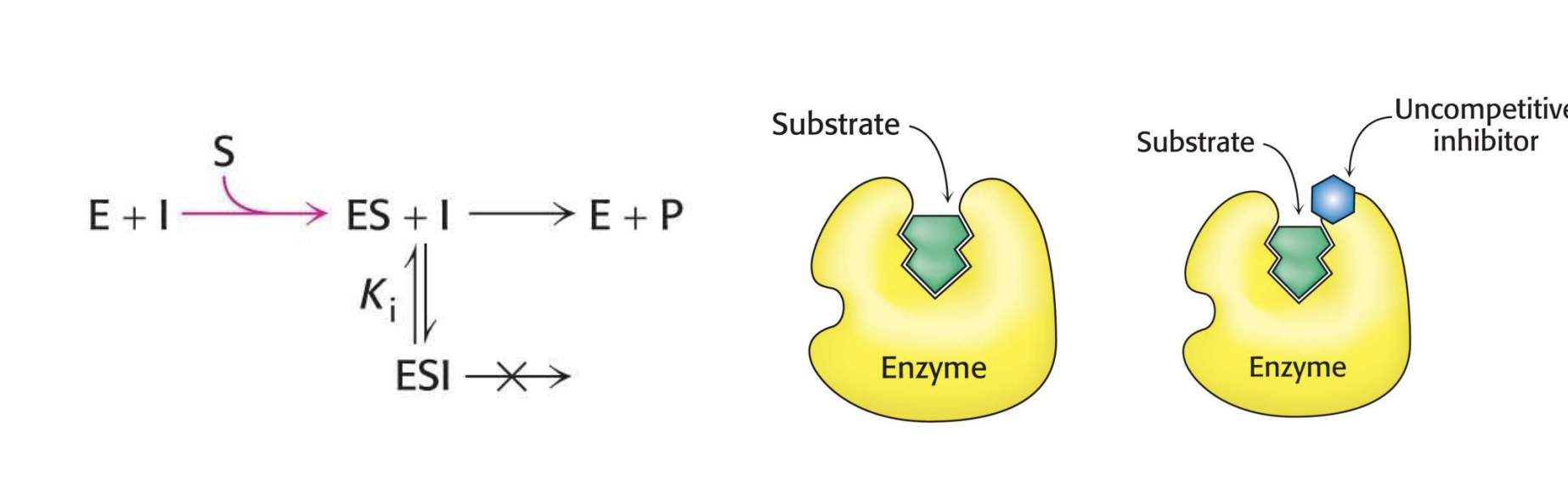
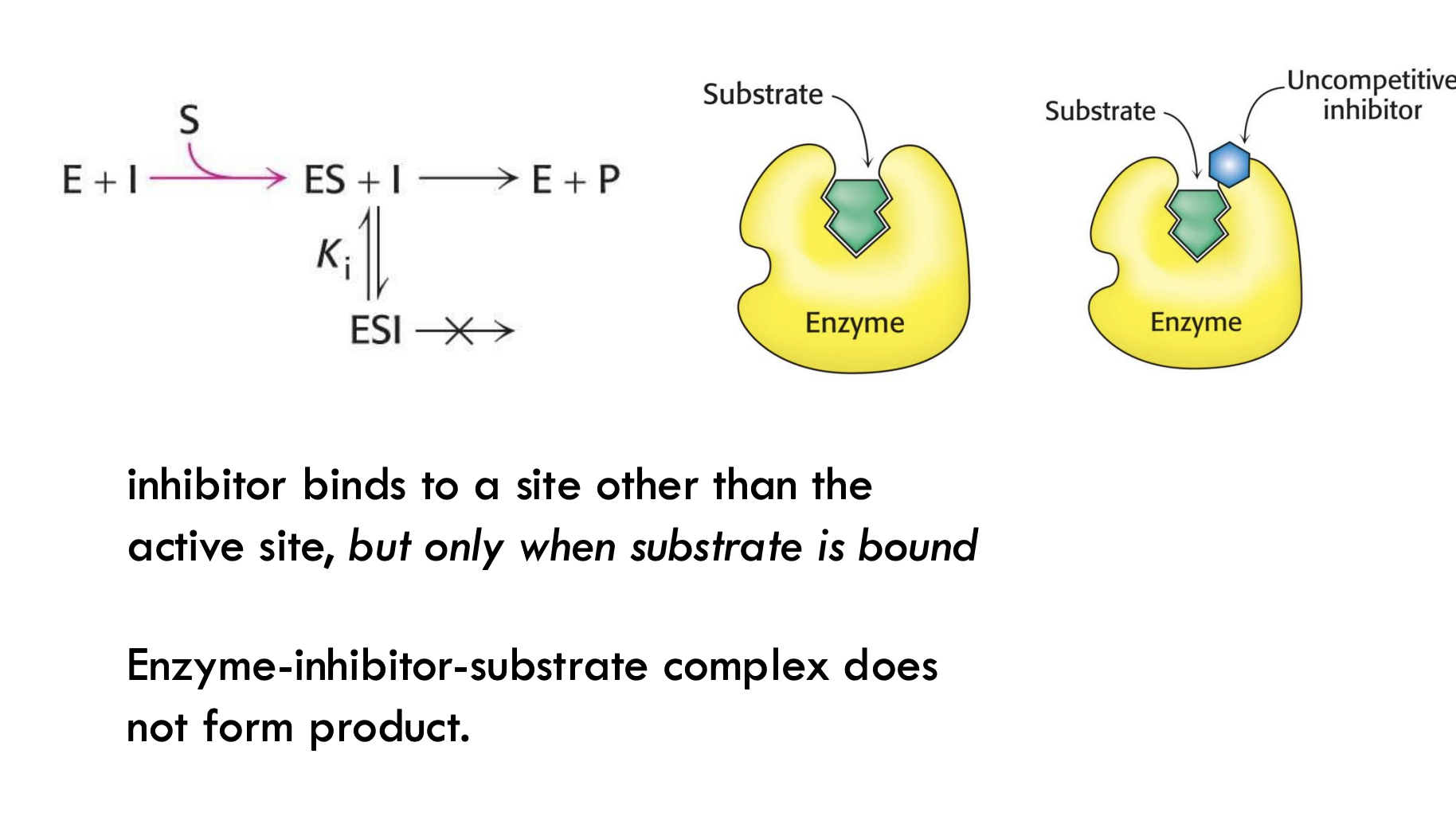
Where does the uncompetitive inhibitor bind, and what does it prevent?
It binds to a site other than the active site and prevents the enzyme-substrate complex from making product.
Simple Example: It's like blocking the exit door of a room after someone (substrate) is already inside — they can’t leave (no product forms).
What happens to VmaxV_{\text{max}}Vmax during uncompetitive inhibition?
Vmax decreases because the inhibitor prevents the enzyme-substrate complex from making product.
Simple Example: It's like jamming the finish line — even if the runners (substrates) start the race, they can’t complete it.
How does uncompetitive inhibition affect KMK_MKM?
Apparent KMK_MKM decreases — the enzyme seems to bind substrate better because the complex is stabilized.
Simple Example: It's like customers getting stuck inside a store — it looks like the store is very "popular" (good binding), but no one is checking out (making product).
Can uncompetitive inhibition be overcome by adding more substrate?
No, adding more substrate does not fix it because the inhibitor only binds when substrate is already bound.
Simple Example: It’s like locking a door after people have already entered — adding more people doesn’t unlock the door.
What happens to VmaxV_{\text{max}}Vmax during noncompetitive inhibition?
Vmax decreases because the enzyme's function is blocked whether or not the substrate is bound.
Simple Example: It's like someone breaking the machine — even if the materials (substrate) are there, the machine can't work.
What happens to KMK_MKM during noncompetitive inhibition?
KM stays the same — substrate can still bind normally, but the enzyme can't do anything after binding.
Simple Example: It's like customers still getting into a broken store — they can enter, but the cash registers don't work, so no sales happen.
Why is it called "noncompetitive" inhibition?
Because the inhibitor does not compete with the substrate for the active site — it binds elsewhere and blocks the reaction no matter what.
Simple Example: It's like sabotaging the store’s power supply instead of blocking the door — people still walk in, but nothing runs inside.
How do competitive, uncompetitive, and noncompetitive inhibition differ?
Competitive: Inhibitor competes for the active site; KMK_MKM increases, VmaxV_{\text{max}}Vmax stays the same.
Example: Like fighting over a seat in a crowded theater.Uncompetitive: Inhibitor binds only to enzyme-substrate complex; both KMK_MKM and VmaxV_{\text{max}}Vmax decrease.
Example: Like locking people inside a ride — they can’t leave.Noncompetitive: Inhibitor binds elsewhere, whether substrate is bound or not; VmaxV_{\text{max}}Vmax decreases, KMK_MKM stays the same.
Example: Like cutting the power — people still enter but nothing works.
What are group-specific reagents in enzyme inhibition?
Group-specific reagents react with specific R-groups (side chains) of amino acids and form a covalent bond, causing irreversible inhibition.
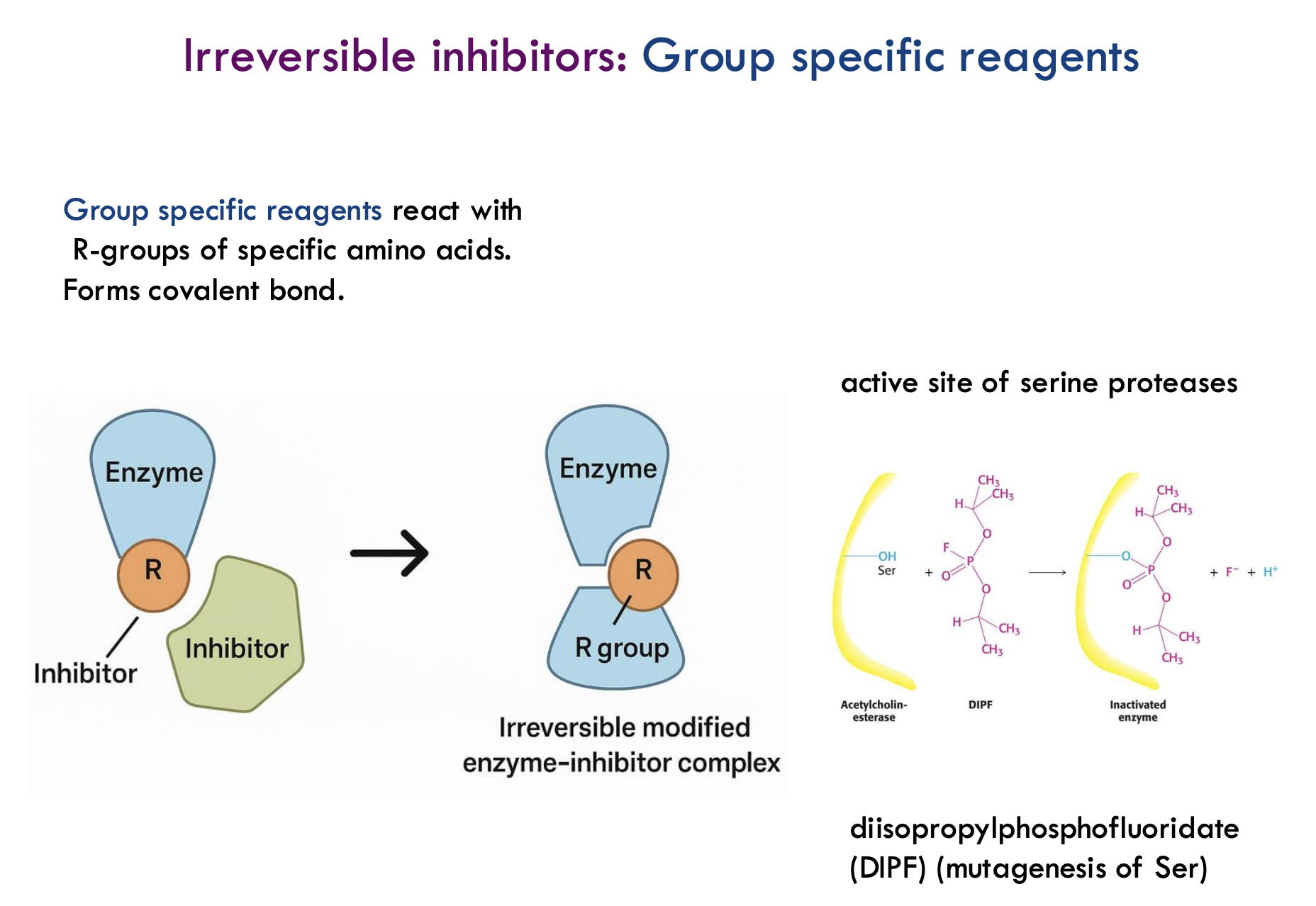
What happens to the enzyme when a group-specific reagent binds?
The enzyme becomes permanently modified and loses its activity — it can no longer perform its normal function.
What is an example of a group-specific reagent and what does it target?
DIPF (diisopropylphosphofluoridate) targets the hydroxyl (-OH) group of serine residues, permanently inactivating enzymes like acetylcholinesterase.
Why are serine residues important targets for group-specific reagents?
Serine residues often play a key catalytic role in enzymes like serine proteases, making them critical for enzyme function.
What is an easy analogy for how irreversible inhibition by group-specific reagents works?
It's like gluing a lock permanently shut — the enzyme (lock) can no longer work or bind anything.
What are affinity labels (substrate analogs)?
Molecules that are structurally similar to the enzyme’s natural substrate but irreversibly inhibit the enzyme by covalently modifying a residue in the active site.
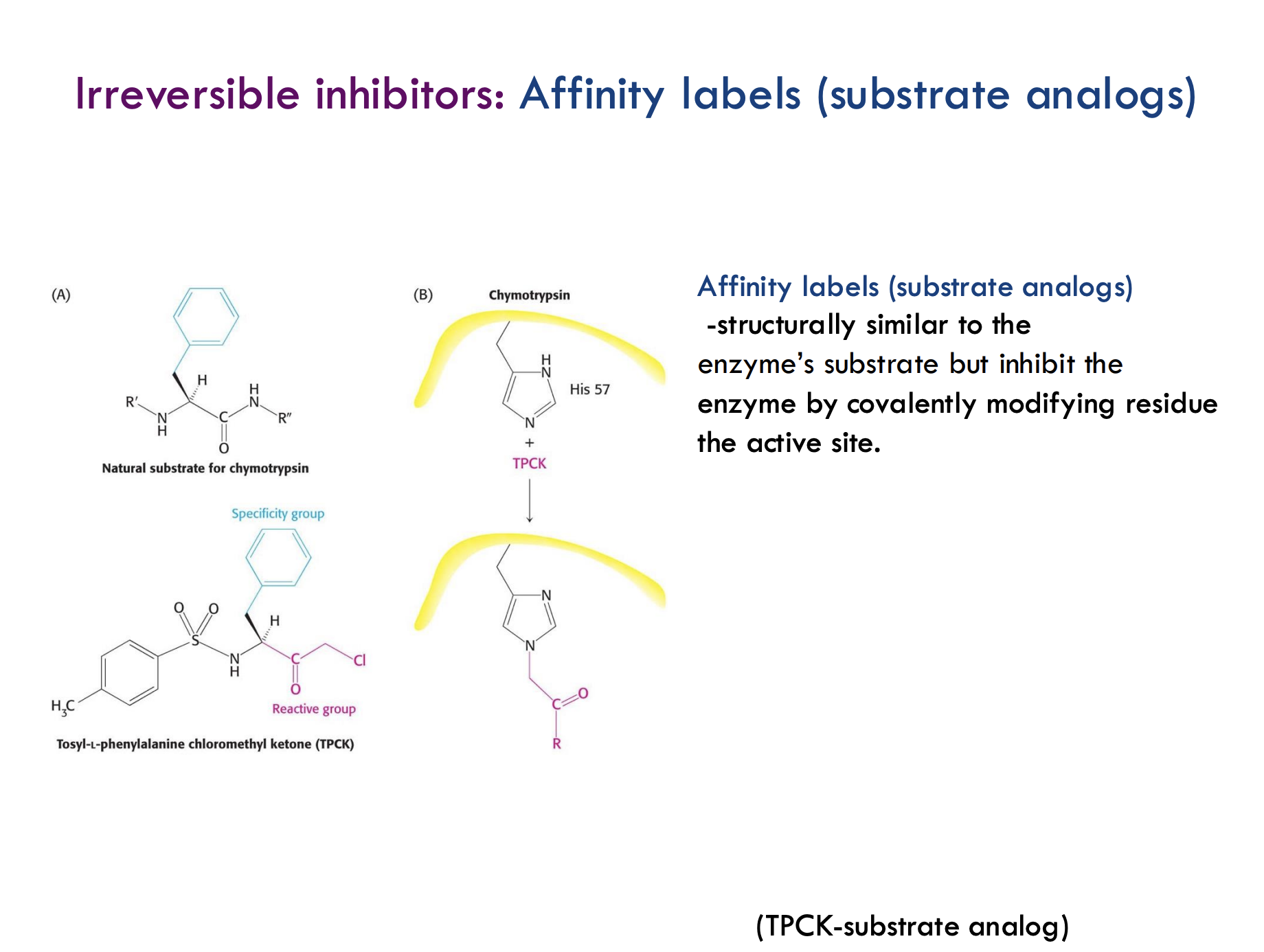
How do affinity labels inhibit enzymes?
They mimic the substrate, bind to the active site, and then form a covalent bond with an active site residue, permanently inactivating the enzyme.
What is an example of an affinity label?
TPCK (tosyl-L-phenylalanine chloromethyl ketone) is an affinity label that targets histidine (His57) in the enzyme chymotrypsin's active site.
What is the "reactive group" and "specificity group" in TPCK?
The specificity group helps TPCK recognize chymotrypsin, and the reactive group forms the covalent bond with His57.
What’s a simple analogy for how affinity labels work?
It’s like a fake key (affinity label) that fits into a lock (enzyme) and then superglues itself inside — permanently breaking the lock.
What are suicide inhibitors (mechanism-based inhibitors)?
They bind to the enzyme like a normal substrate, but during catalysis, the enzyme modifies them into a reactive form that irreversibly inactivates the enzyme.
How do suicide inhibitors work?
The enzyme accidentally "activates" the inhibitor into a reactive intermediate that permanently blocks the enzyme.
What’s a famous example of a suicide inhibitor?
Penicillin — it binds to the enzyme glycopeptide transpeptidase and forms a stable, inactive penicilloyl-enzyme complex.
What are transition-state analogs?
They are inhibitors that mimic the transition state of a reaction, but cannot actually be processed by the enzyme.
How do transition-state analogs work?
They fit perfectly into the enzyme’s active site (like the real transition state would), but get stuck because they can’t react — blocking the enzyme non-covalently.
What are the four main types of catalytic mechanisms enzymes use?
Acid-base catalysis: Enzyme donates or accepts protons.
Covalent catalysis: Enzyme forms a covalent bond with the substrate.
Metal ion catalysis: Metal ions help stabilize charges or activate water.
Catalysis by approximation/orientation: Enzyme brings molecules close together and aligns them perfectly for reaction.
What is acid-base catalysis and why is it important?
Acid-base catalysis is the transfer of a proton (H⁺) from one molecule to another.
It stabilizes charged intermediates during a reaction.
It can be specific (using water) or general (using amino acid side chains like His, Glu).
Example:
Histidine in an enzyme can grab a proton to stabilize a negative charge forming during the reaction.
Which amino acids commonly participate in acid-base catalysis, and how?
General acid (proton donor): Glu, Asp, Lys, Arg, His, Cys, Ser, Tyr.
General base (proton acceptor): Their deprotonated forms (like COO⁻, NH₂, etc.).
Example:
Glu (glutamate) can donate a proton (acid) and stabilize a reaction intermediate.
What is the difference between specific and general acid-base catalysis?
Specific catalysis: Proton transfer directly from water (H₃O⁺ or OH⁻).
General catalysis: Proton transfer involving amino acid side chains (like His, Glu) acting as proton donors or acceptors.
Example:
Specific: Proton from water helps in a reaction.
General: Histidine side chain donates/accepts a proton during the reaction.
What is covalent catalysis?
Covalent catalysis is when a temporary covalent bond forms between the enzyme and substrate to speed up the reaction.
Example:
The enzyme lends a part of itself (like a functional group) to help break or form bonds more easily.
How does covalent catalysis make a reaction faster?
It creates a reactive intermediate that lowers activation energy, making it easier for the reaction to proceed.
Extra:
The enzyme returns to its original state at the end — it’s not permanently changed!
What is metal ion catalysis?
Metal ion catalysis is when an enzyme uses a metal ion to help in the reaction by stabilizing negative charges, transferring electrons, or activating water.
Example:
A metal ion like Mg²⁺ helps pull electrons away from water, making it easier to break bonds.
What are four key ways a metal ion assists catalysis?
Orients the substrate properly for the reaction
Makes water more active (easier to break bonds)
Stabilizes negative charges during the reaction
Helps transfer electrons
Tip:
Think of the metal as a "helper magnet" that pulls things into the right place!
What is catalysis by approximation and orientation?
It’s when the enzyme speeds up the reaction by bringing substrates close together (approximation) and lining them up correctly (orientation) so they react faster.
Example:
Imagine two puzzle pieces — the enzyme holds and rotates them so they fit together perfectly!
What are three ways enzymes help in approximation/orientation catalysis?
Hold substrates in place
Restrict movement (so they don’t wiggle around)
Align reactive groups properly for the reaction to happen
Quick Tip:
Think of the enzyme as a coach positioning players perfectly before a big play!
What is the function of Chymotrypsin during digestion?
Chymotrypsin is a serine protease that breaks down dietary proteins into small peptides by cutting specific peptide bonds on the peptide backbone (hydrolysis).
Example:
It acts like scissors that only cut at certain spots in a protein chain!
Where exactly does Chymotrypsin cut the peptide chain?
Chymotrypsin cleaves the peptide bond adjacent to aromatic amino acids (like phenylalanine, tyrosine, tryptophan), and speeds up the reaction by a factor of 10⁹!
Example:
It looks for a "fancy ring" (aromatic group) in the protein and cuts right next to it.
What is a zymogen?
A zymogen is an inactive precursor of an enzyme that needs to be cleaved to become active.
Example:
Chymotrypsinogen must be cut to become active chymotrypsin!
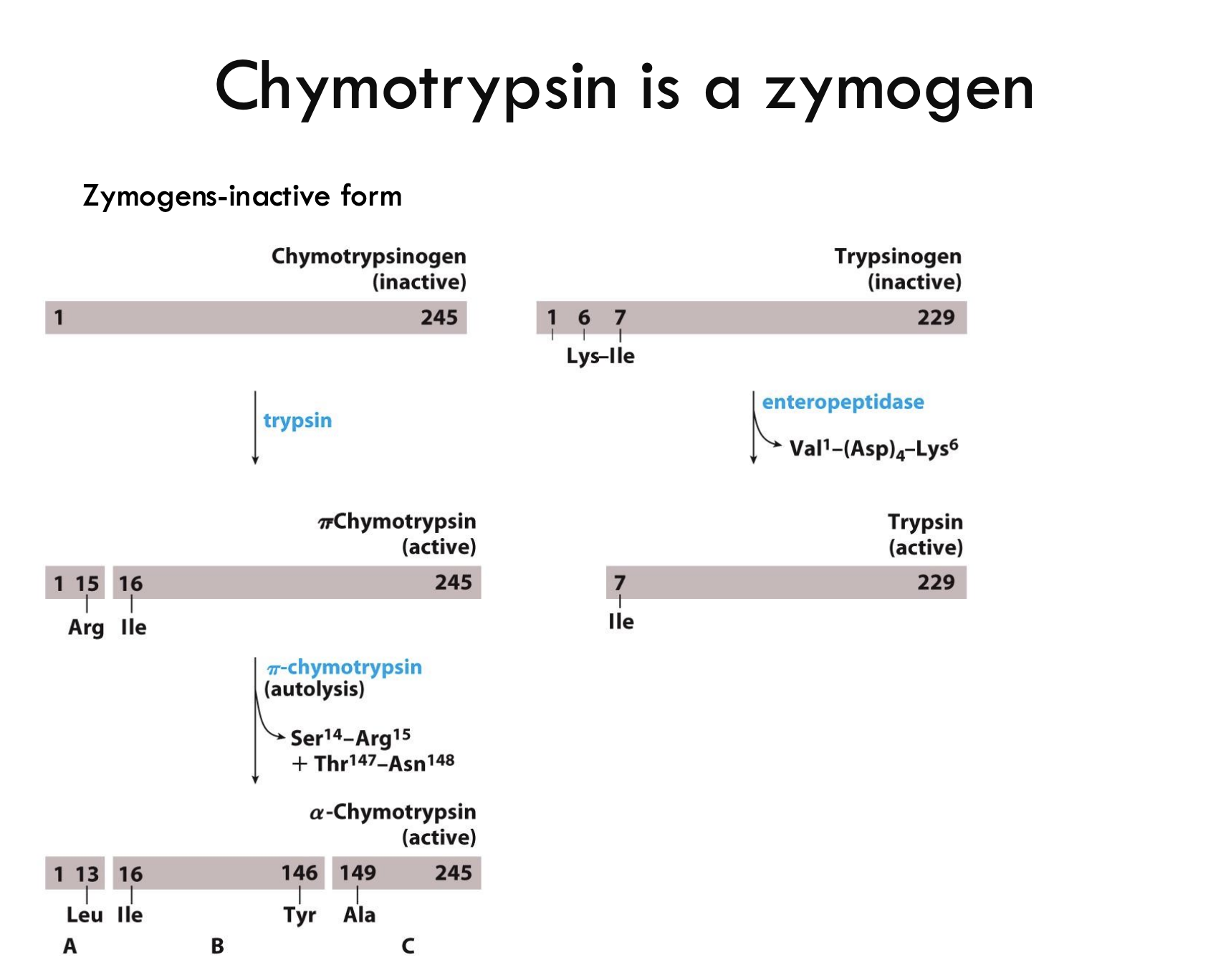
How is chymotrypsinogen activated?
Trypsin cleaves chymotrypsinogen between Arg15 and Ile16, forming π-chymotrypsin (partially active form).
Example:
Think of Trypsin like a "key" unlocking chymotrypsin's potential!
What additional changes convert π-chymotrypsin into fully active α-chymotrypsin?
π-chymotrypsin self-digests (autolysis) at two spots:
Ser14-Arg15
Thr147-Asn148
creating three chains (A, B, C) held by disulfide bonds.
Example:
π-chymotrypsin trims itself to become the final mature enzyme!
What enzyme activates trypsinogen into trypsin?
Enteropeptidase cleaves trypsinogen at the bond between Val-(Asp)₄-Lys⁶, activating it into trypsin.
Example:
Enteropeptidase = the "starter" that kicks off the whole digestive enzyme cascade!
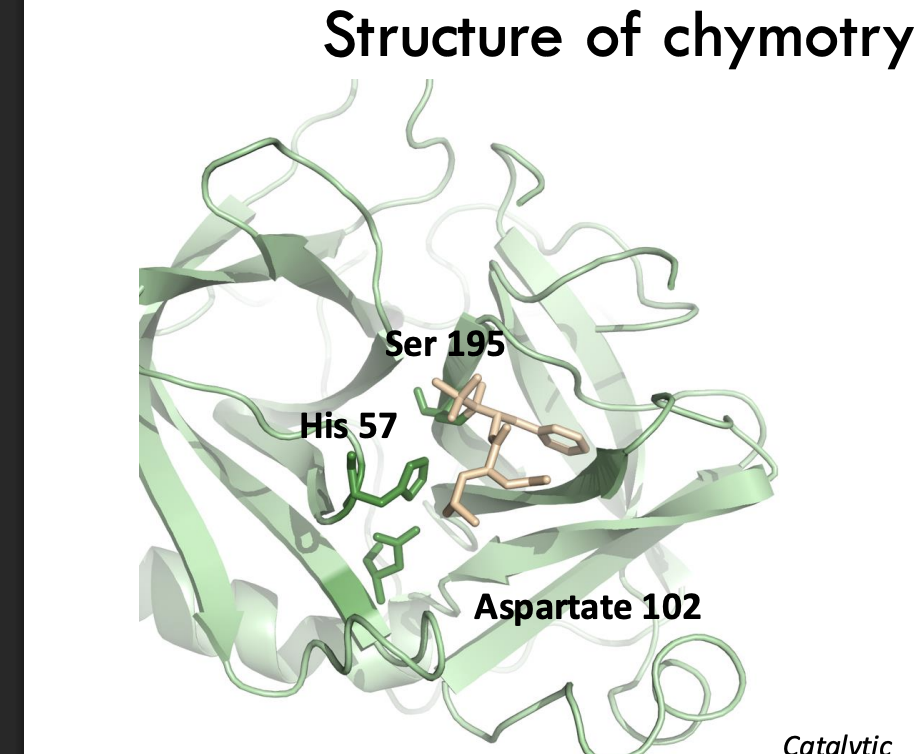
What are the three key residues in the chymotrypsin active site and what is their role?
Ser195 – Acts as the "knife" (nucleophile) that attacks the peptide bond.
His57 – Acts as a "relay" to transfer protons.
Asp102 – Stabilizes His57 by holding it in place.
Example:
Think of Ser-His-Asp as a knife-sharpener-holder team.
What is the function of the S1 pocket in chymotrypsin?
The S1 pocket is a hydrophobic binding site that positions aromatic side chains (like phenylalanine) next to the catalytic triad for cleavage.
Example:
The S1 pocket is like a custom glove that grabs aromatic amino acids tightly.
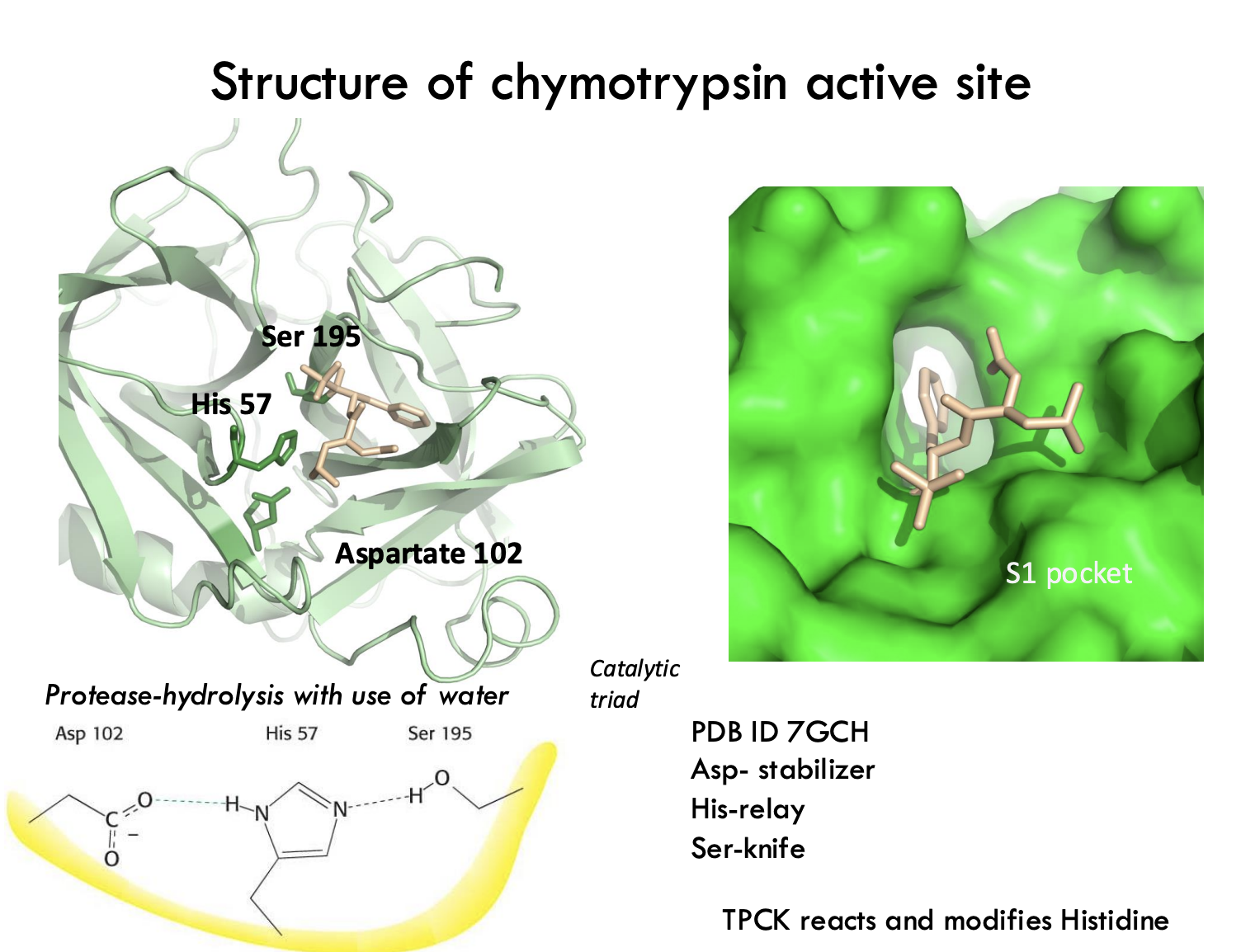
What does TPCK do in chymotrypsin studies?
TPCK is a substrate analog that covalently modifies His57 to identify active site residues.
Example:
TPCK is like fluorescent paint that marks exactly where His57 is!
What is the first step in peptide hydrolysis by chymotrypsin?
Histidine acts as a general base, accepting a proton from serine, creating a reactive nucleophilic ion.
Example:
His "steals" a proton from Ser so Ser can attack the peptide bond like a sword.
What does the serine nucleophile do after activation?
The activated serine attacks the carbonyl carbon of the peptide bond, forming a tetrahedral intermediate.
Example:
Think of Ser acting like a ninja striking the carbonyl.
What role does the oxyanion hole play during chymotrypsin catalysis?
The oxyanion hole stabilizes the negative charge that forms on the oxygen atom during the tetrahedral intermediate.
Example:
The oxyanion hole is like a comfy chair that "catches" and stabilizes the negative oxygen.
How is substrate specificity achieved in chymotrypsin?
The substrate's hydrophobic side chain fits snugly into a hydrophobic pocket near the active site.
Example:
It’s like a puzzle piece snapping perfectly into a hydrophobic slot.
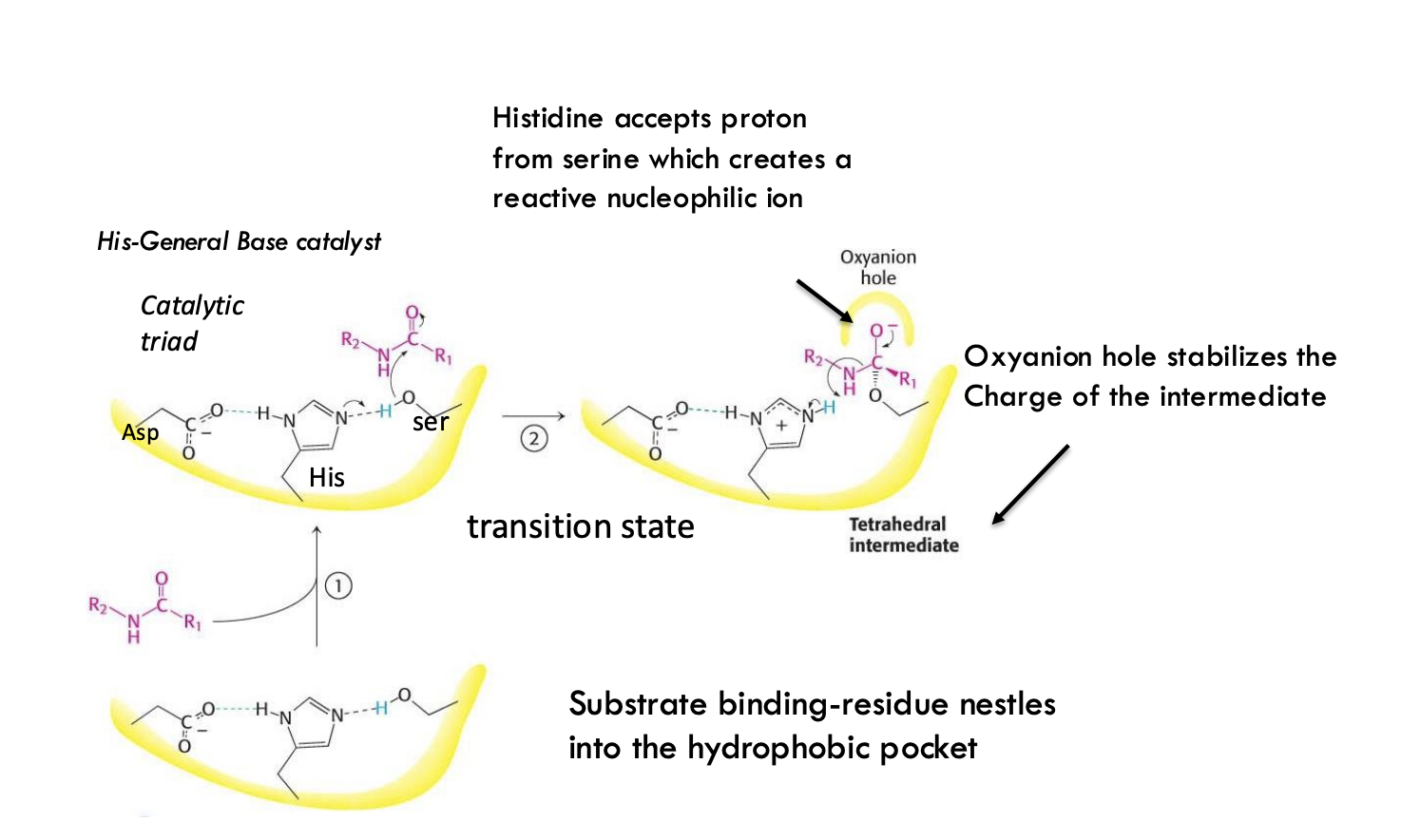
What important intermediate is formed during chymotrypsin peptide bond cleavage?
A tetrahedral intermediate is formed, stabilized by the oxyanion hole, before collapsing to cleave the peptide bond.
Example:
Picture the transition state as a wobbly 3D pyramid shape that needs stabilizing.
What happens after the first tetrahedral intermediate collapses in chymotrypsin catalysis?
The tetrahedral intermediate collapses, transferring a proton to the leaving group, creating an acyl-enzyme intermediate.
Example:
Think of the intermediate "falling apart" to hand off part of itself and make an acyl-enzyme.
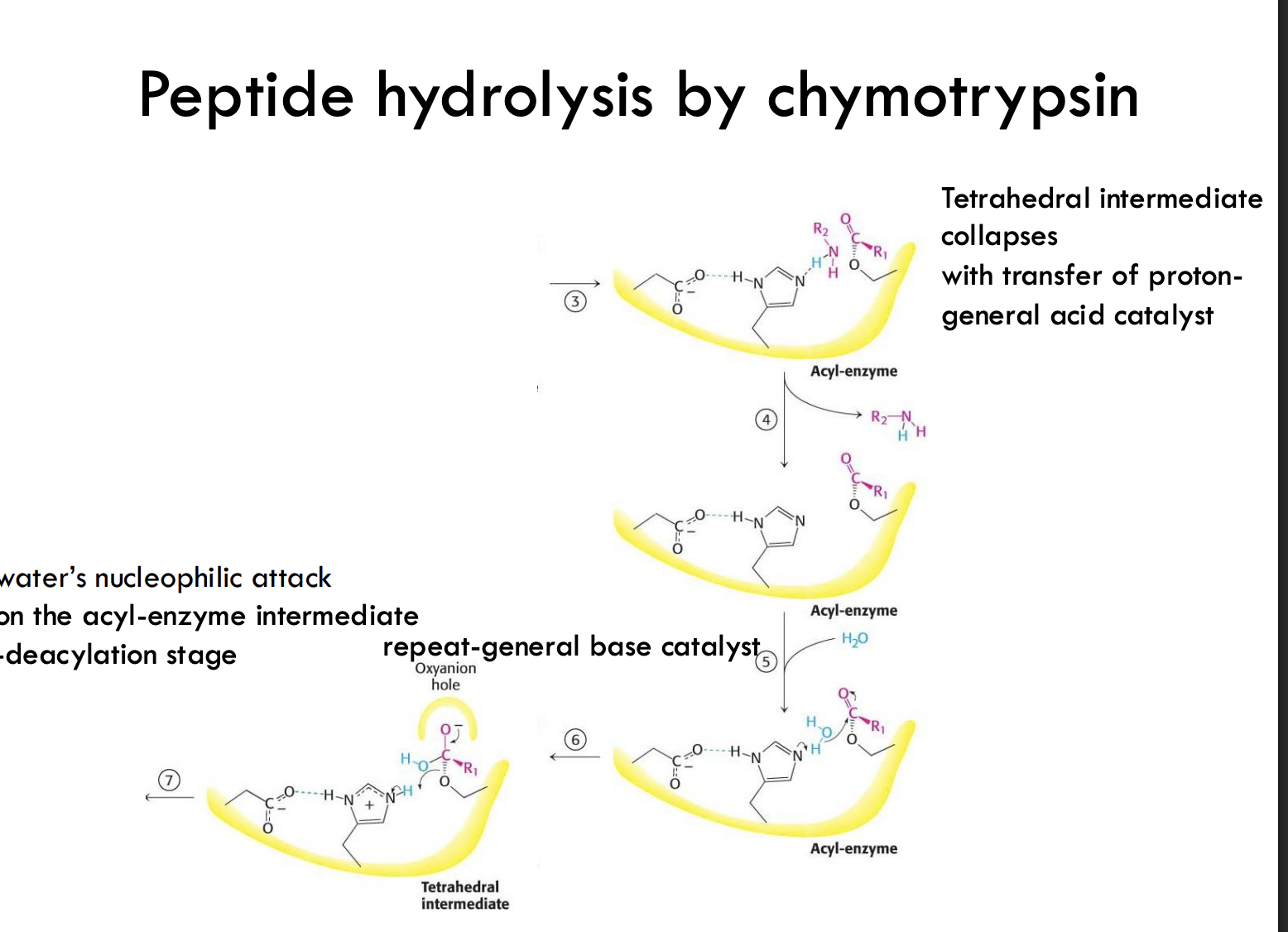
What does the acyl-enzyme intermediate represent?
It's a covalently bound intermediate where the substrate's carbonyl group is temporarily attached to the enzyme's serine.
Example:
Like Velcro: the broken piece sticks temporarily to the enzyme.
What is the role of water in the second half of the chymotrypsin reaction?
Water attacks the acyl-enzyme intermediate as a nucleophile, leading to hydrolysis and breaking the bond.
Example:
Water acts like a second ninja, sneaking in to finish cutting the Velcro bond.
How does the enzyme facilitate water's nucleophilic attack?
Histidine acts again as a general base, pulling a proton off water to make it a better nucleophile.
Example:
His acts like a coach, prepping water to hit harder!
What happens after the second tetrahedral intermediate forms?
t collapses, releasing the final peptide product and regenerating the free enzyme.
Example:
The enzyme spits out the final product and is ready to start a new mission.
What are the two main stages of peptide bond hydrolysis by chymotrypsin, and what happens in each?
Stage 1 (Acylation):
Ser195 attacks the peptide bond.
A tetrahedral intermediate forms and collapses.
The C-terminal fragment leaves, and an acyl-enzyme intermediate forms (substrate stuck to Ser).
Stage 2 (Deacylation):
Water attacks the acyl-enzyme.
A second tetrahedral intermediate forms and collapses.
The N-terminal fragment is released, and the enzyme is reset.
Simple Example:
Stage 1 = Knife stabs and holds half the sandwich.
Stage 2 = Water sprays, enzyme drops sandwich pieces, ready for the next bite.
What is a common functional theme in proteins regarding molecule binding?
Many proteins use reversible binding of molecules, like protein-ligand or enzyme-substrate interactions.
How are protein binding sites designed for their ligands?
Binding sites are complementary to ligand size, shape, charge, hydrophobicity, and hydrophilicity, ensuring specific interaction—similar to enzyme active sites.
How does protein flexibility support function?
Proteins can adapt structurally to ligands (induced fit) through noncovalent interactions, preserving protein structure.
How do multimeric (multi-subunit) proteins behave during binding?
In multisubunit proteins, binding or conformational change in one subunit can affect the conformation of neighboring subunits.
Why can't oxygen (O₂) just float around freely in biological systems?
O₂ has low solubility in solution and protein side chains don't bind it well.
Why can't we just use free transition metals to bind O₂?
Free metals can bind O₂ but create dangerous free radicals that can damage cells.
What is the danger of free heme (Fe²⁺) in solution?
Free heme can be oxidized to Fe³⁺, which is highly reactive and harmful.
How does biology solve the problem of oxygen binding?
By using heme that is protein-bound to safely capture oxygen.
What proteins bind oxygen using a protein-bound heme?
Myoglobin (storage) and Hemoglobin (transport).
What is the basic structure of a porphyrin ring?
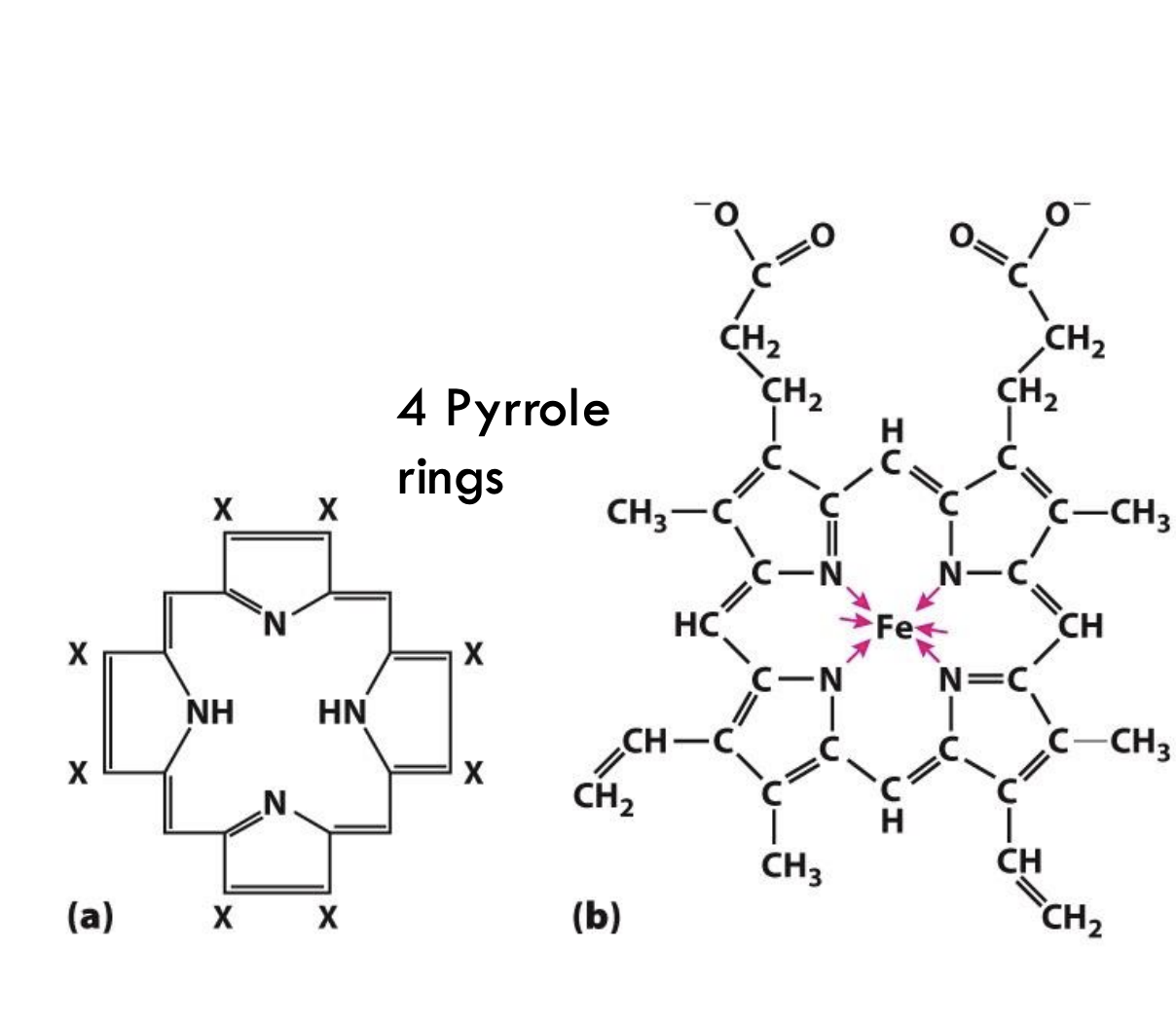
A porphyrin ring is made of four connected pyrrole rings (five-membered rings containing nitrogen atoms).
What makes a porphyrin ring into heme?
When a ferrous ion (Fe²⁺) binds at the center of the porphyrin ring, it forms heme, allowing it to bind oxygen.
What is the prosthetic group in oxygen-binding proteins like myoglobin and hemoglobin?
The prosthetic group is heme (Fe-protoporphyrin IX), where Fe²⁺ binds oxygen.
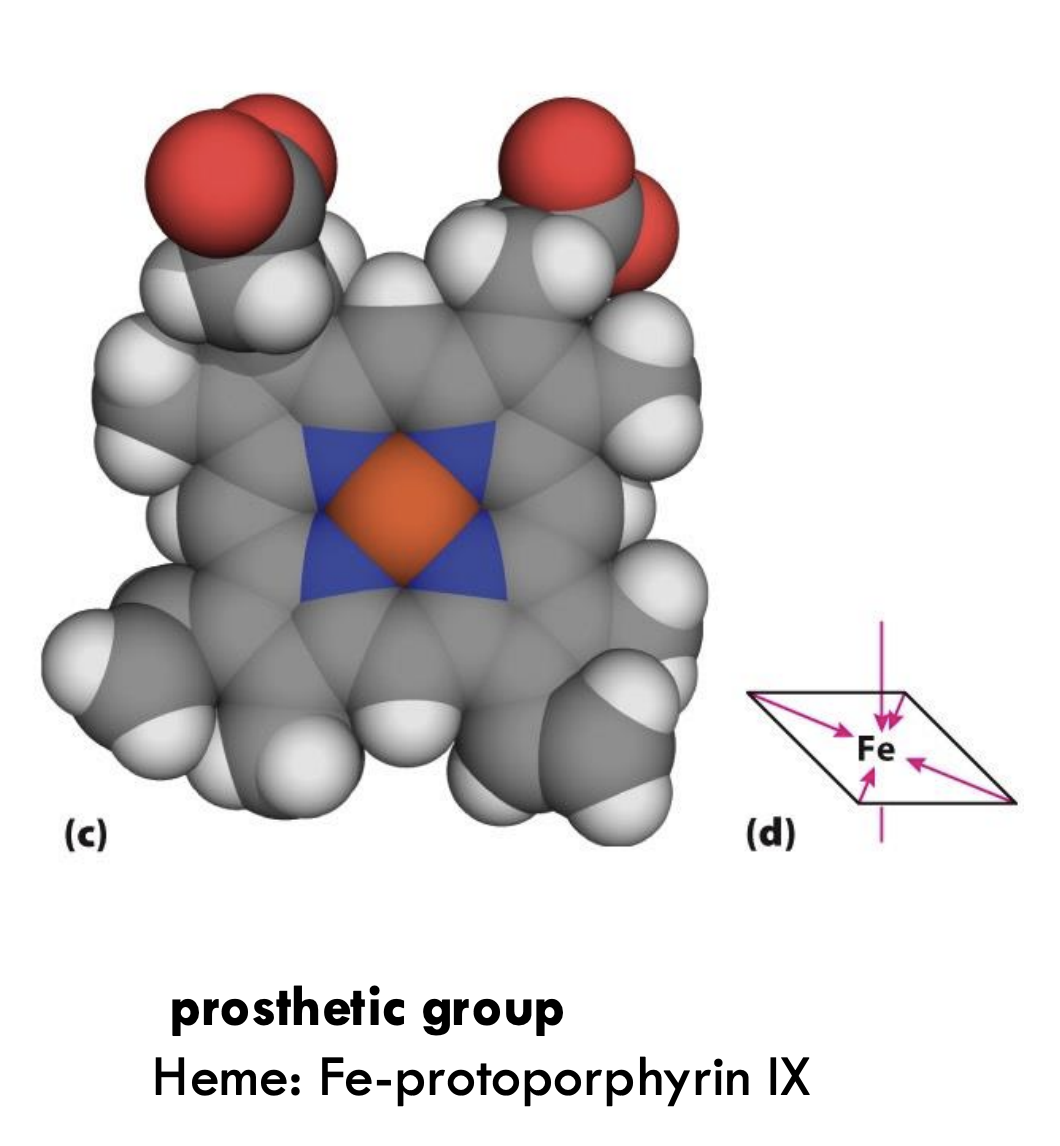
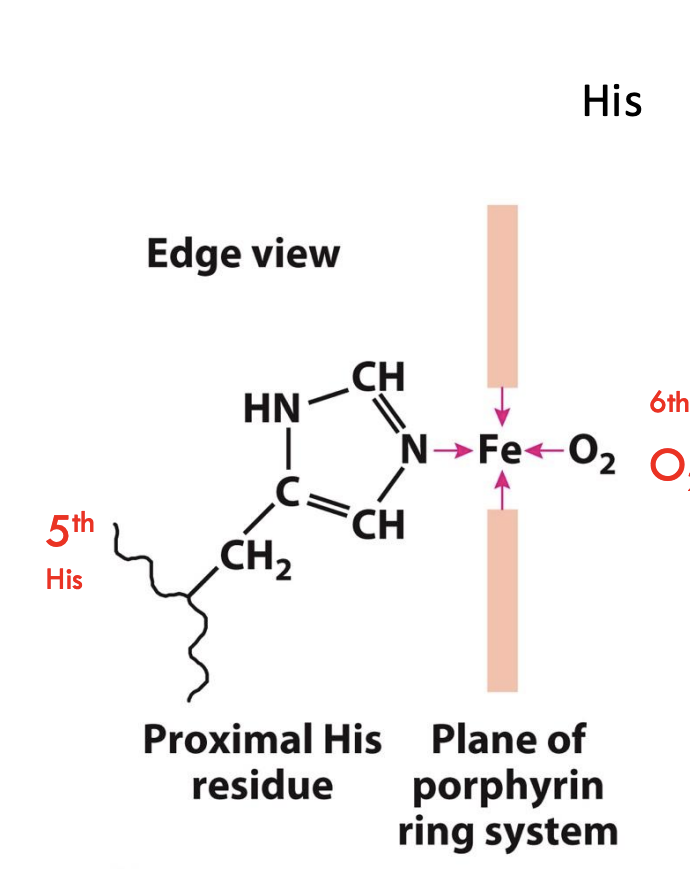
What important residues anchor the heme group in myoglobin?
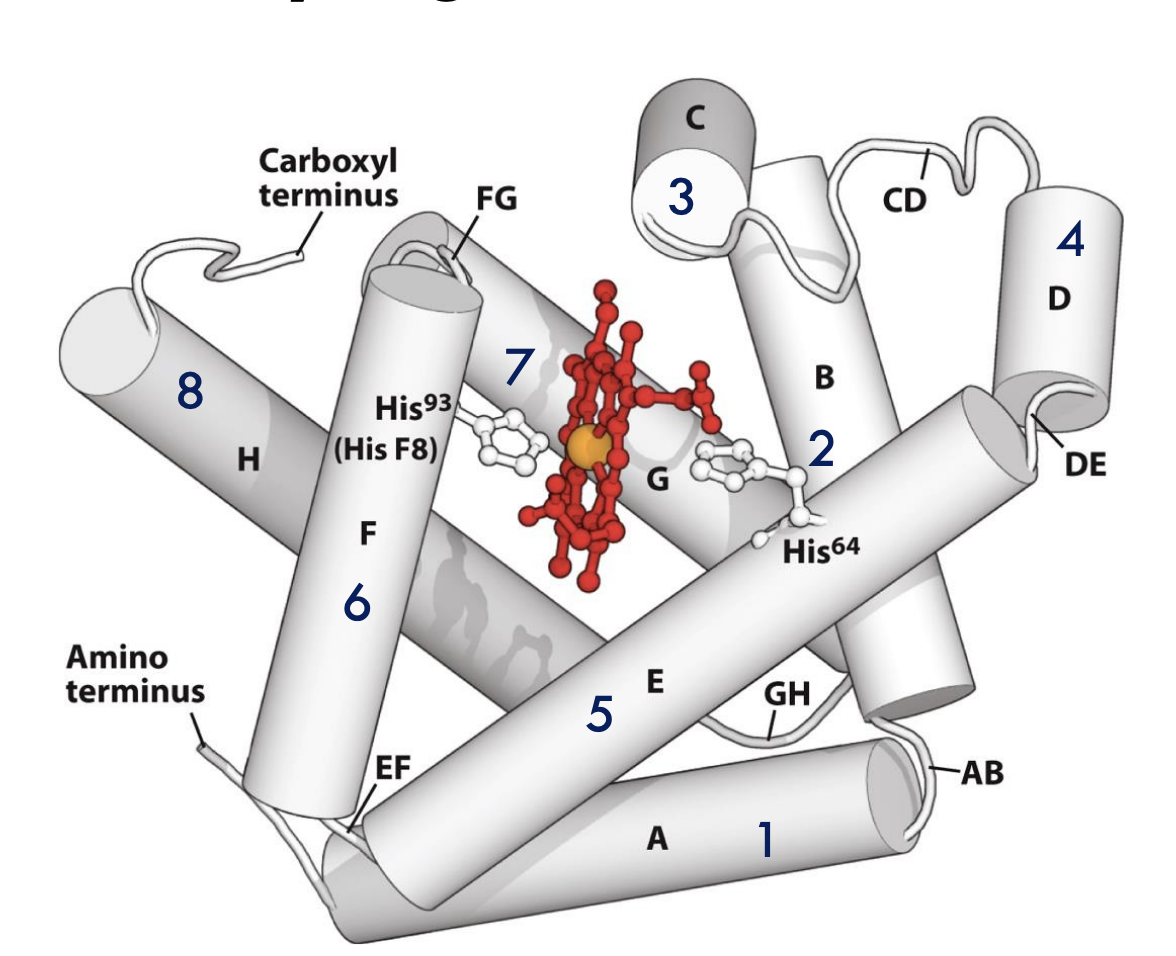
The proximal histidine (His93 or His F8) binds directly to the iron atom of heme, while the distal histidine (His64 or His E7) helps stabilize the bound oxygen molecule.
Who first solved the structure of myoglobin, and what forms were studied?
Perutz and Kendrew (1958) solved the structures of deoxymyoglobin (no oxygen) and oxymyoglobin (with oxygen).
How does the distal histidine (His E7) in myoglobin help oxygen binding?
The distal histidine forms a hydrogen bond with the bound O₂, stabilizing the Fe-O₂ complex and making oxygen binding more secure.
Why does myoglobin reduce carbon monoxide (CO) binding?
Myoglobin's distal histidine (His E7) and the shape of the binding pocket block CO from binding tightly, favoring oxygen (O₂) instead.
What is "molecular breathing" in myoglobin?
"Molecular breathing" refers to the slight opening and closing of the protein structure to allow oxygen to enter and leave the heme pocket.
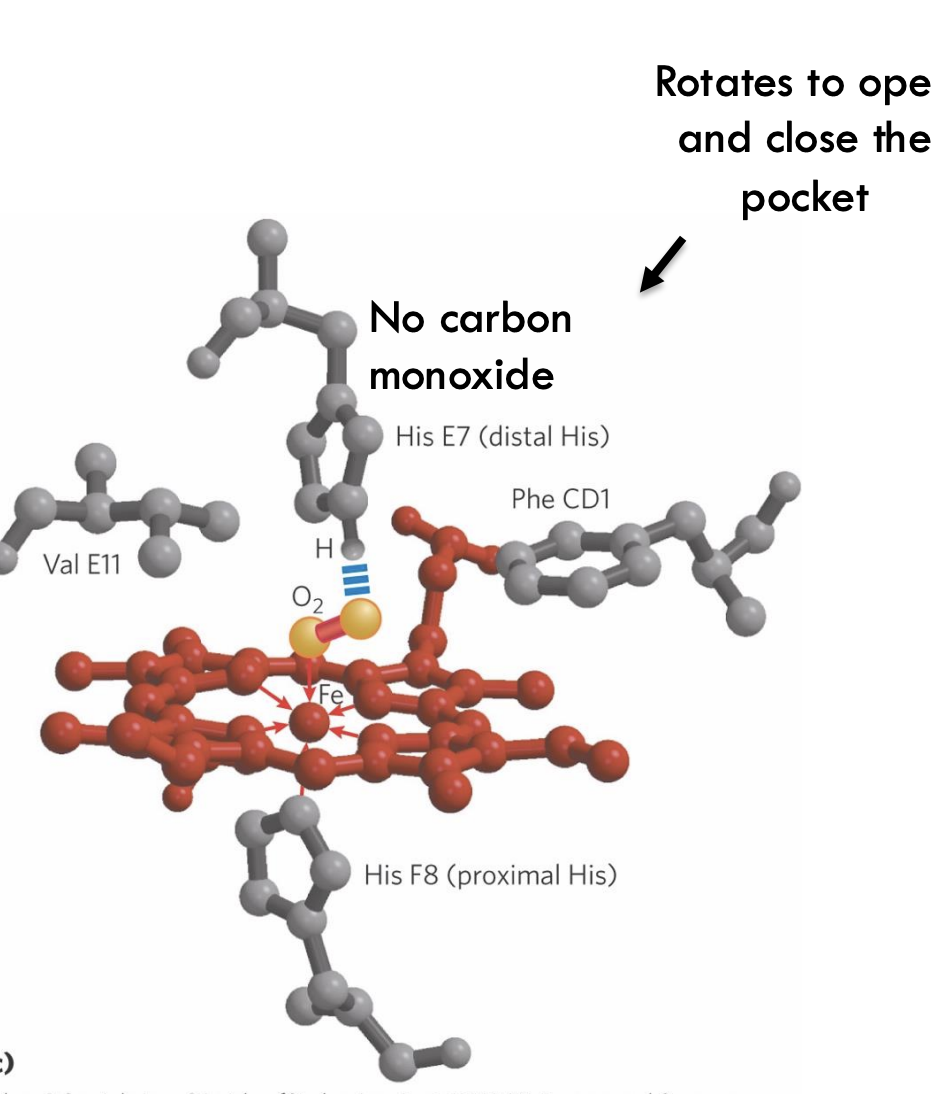
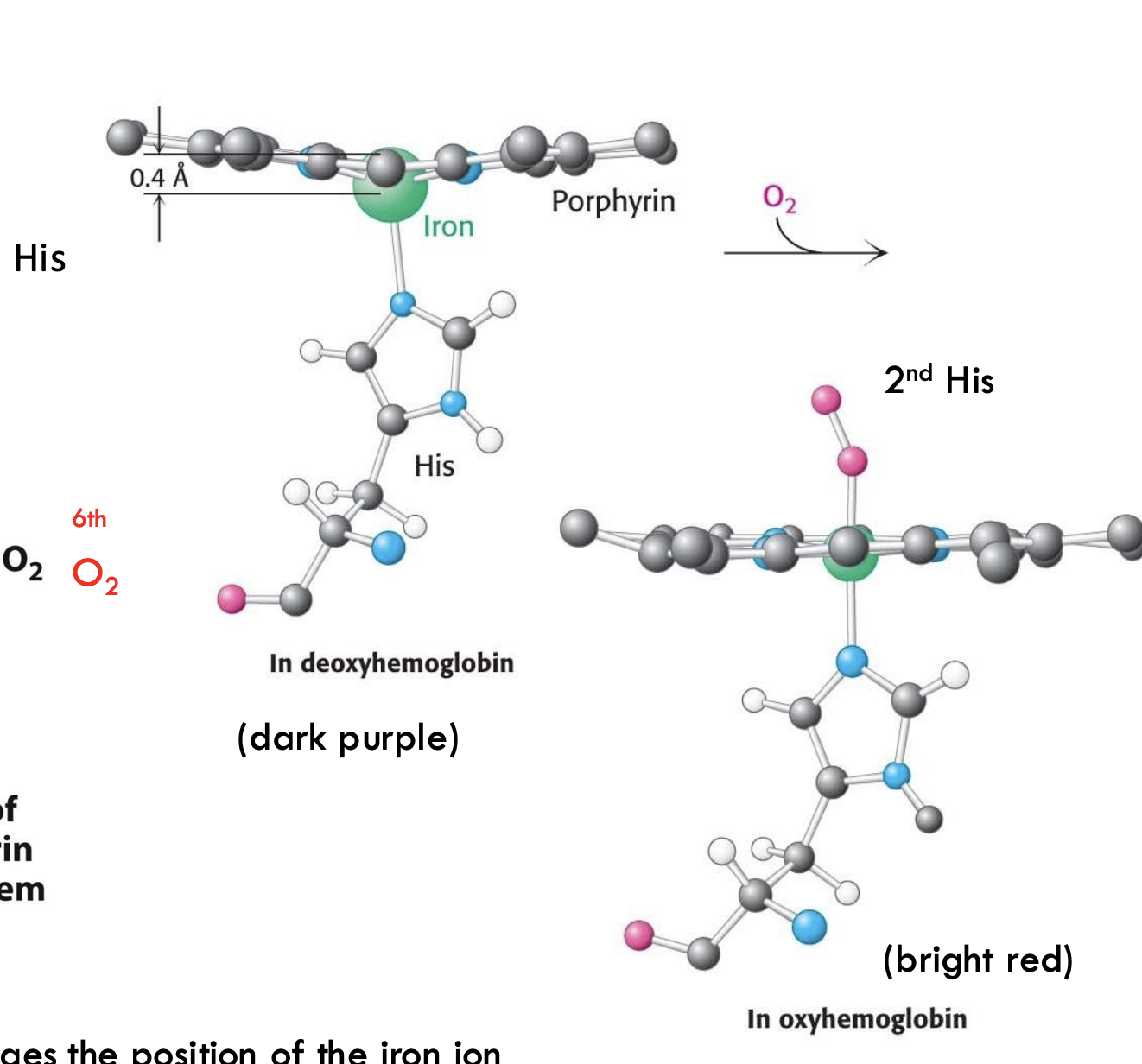
What happens to iron (Fe²⁺) in hemoglobin when oxygen binds?
When O₂ binds, Fe²⁺ moves into the plane of the porphyrin ring, pulling the attached proximal histidine (His F8) with it and triggering a structural change.
How does oxygen binding change the shape of hemoglobin?
Oxygen binding flattens the porphyrin ring, moving iron and the attached His F8, which then shifts the entire hemoglobin subunit, helping neighboring subunits bind oxygen more easily (cooperative binding).
What role does the 5th histidine (His F8) play in hemoglobin?
The 5th histidine (proximal His) binds directly to Fe²⁺, and its movement after oxygen binding helps transmit the conformational change across the protein.
What is the structural relationship between myoglobin and hemoglobin?
Hemoglobin is a tetramer (α₂β₂) made of four subunits, and each subunit is structurally similar to myoglobin, w
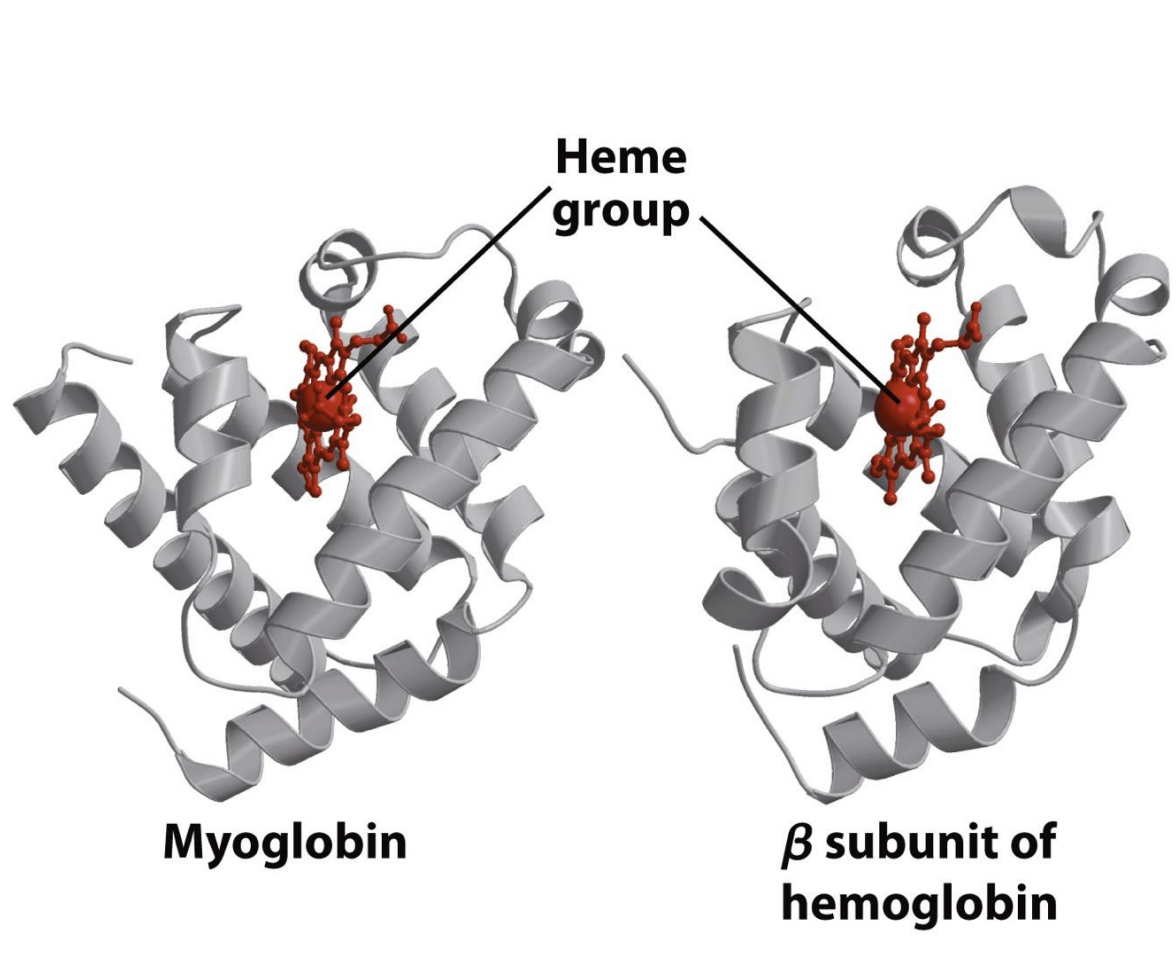
hich is a single polypeptide.
What key component is found in both myoglobin and hemoglobin?
Both myoglobin and hemoglobin contain a heme group that binds oxygen.
How are the amino acid sequences of myoglobin and hemoglobin related?
Their sequences are different, but their three-dimensional structures are very similar because critical residues (like the proximal and distal histidines) are conserved.
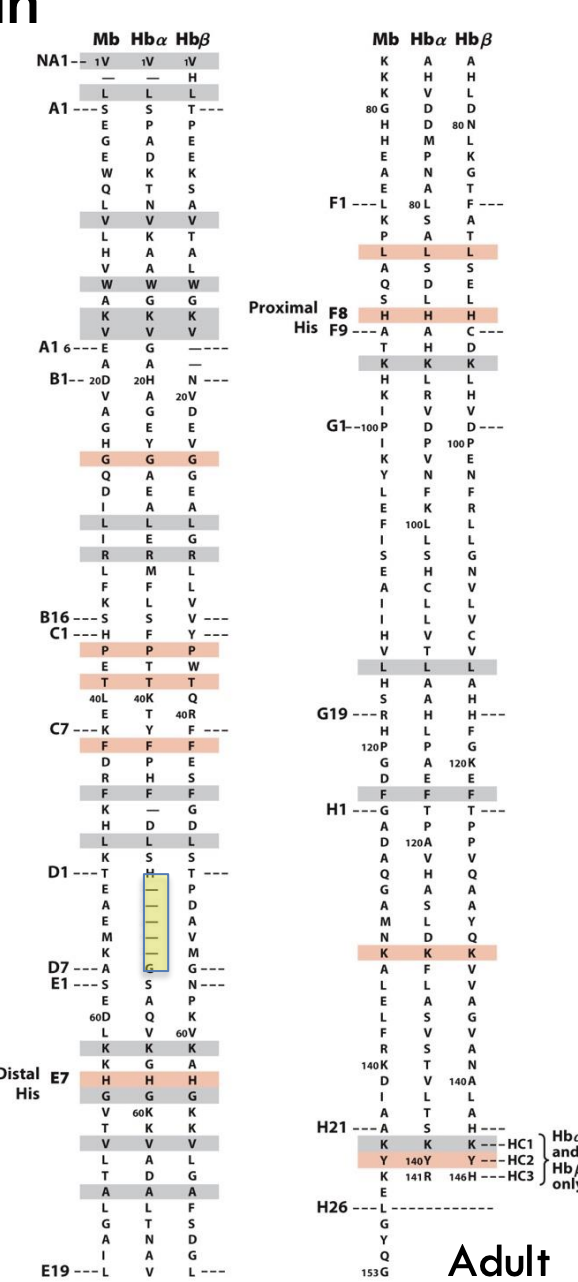
What does it mean when globin proteins show low sequence similarity but high structural similarity?
Even though the amino acid sequences of globin proteins are very different (low sequence similarity), their 3D structures are very similar (high structural similarity). This means that evolution changed the sequence over time but preserved the important helices and the heme pocket, allowing the proteins to keep their shape
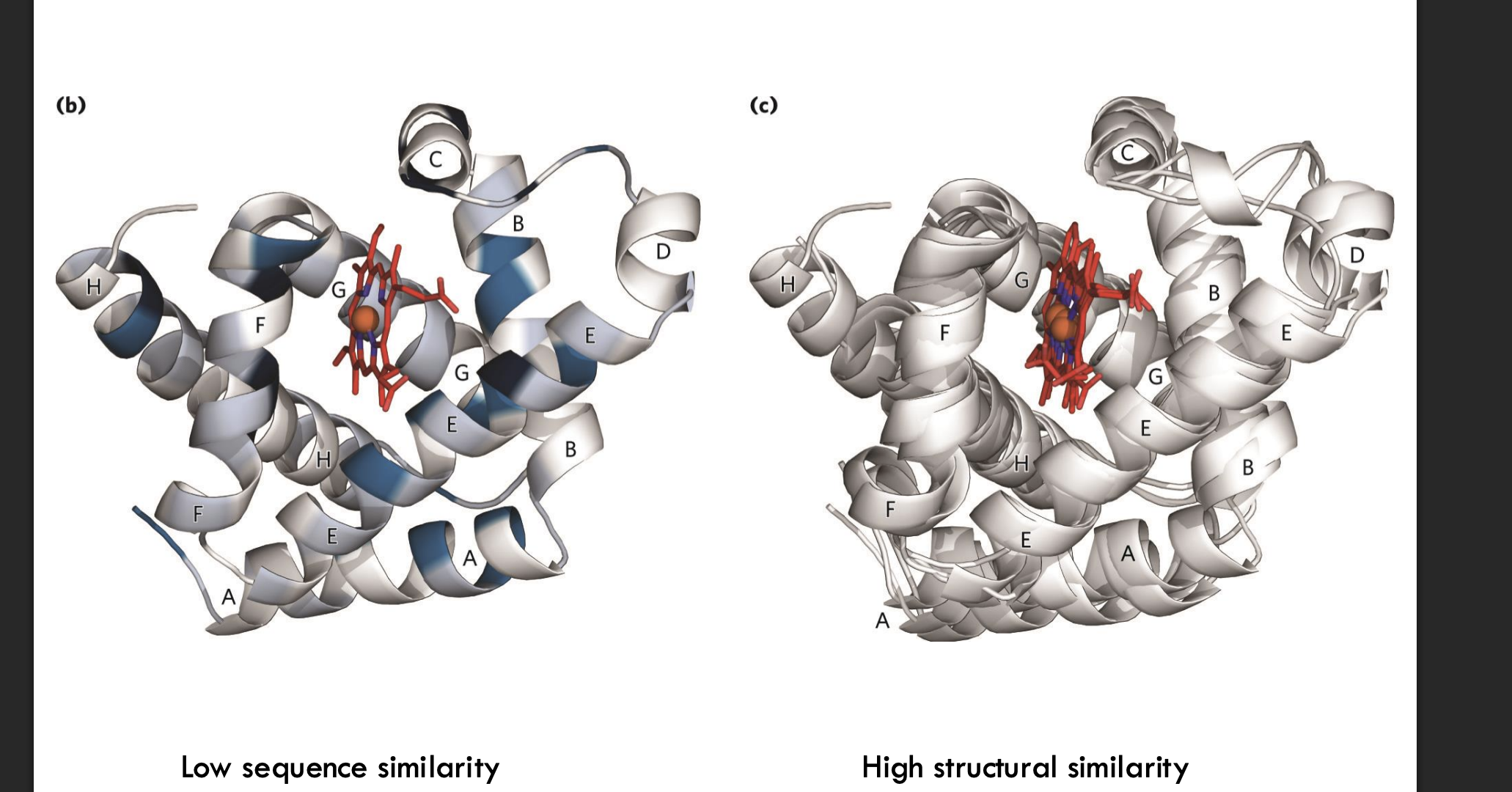
and function.
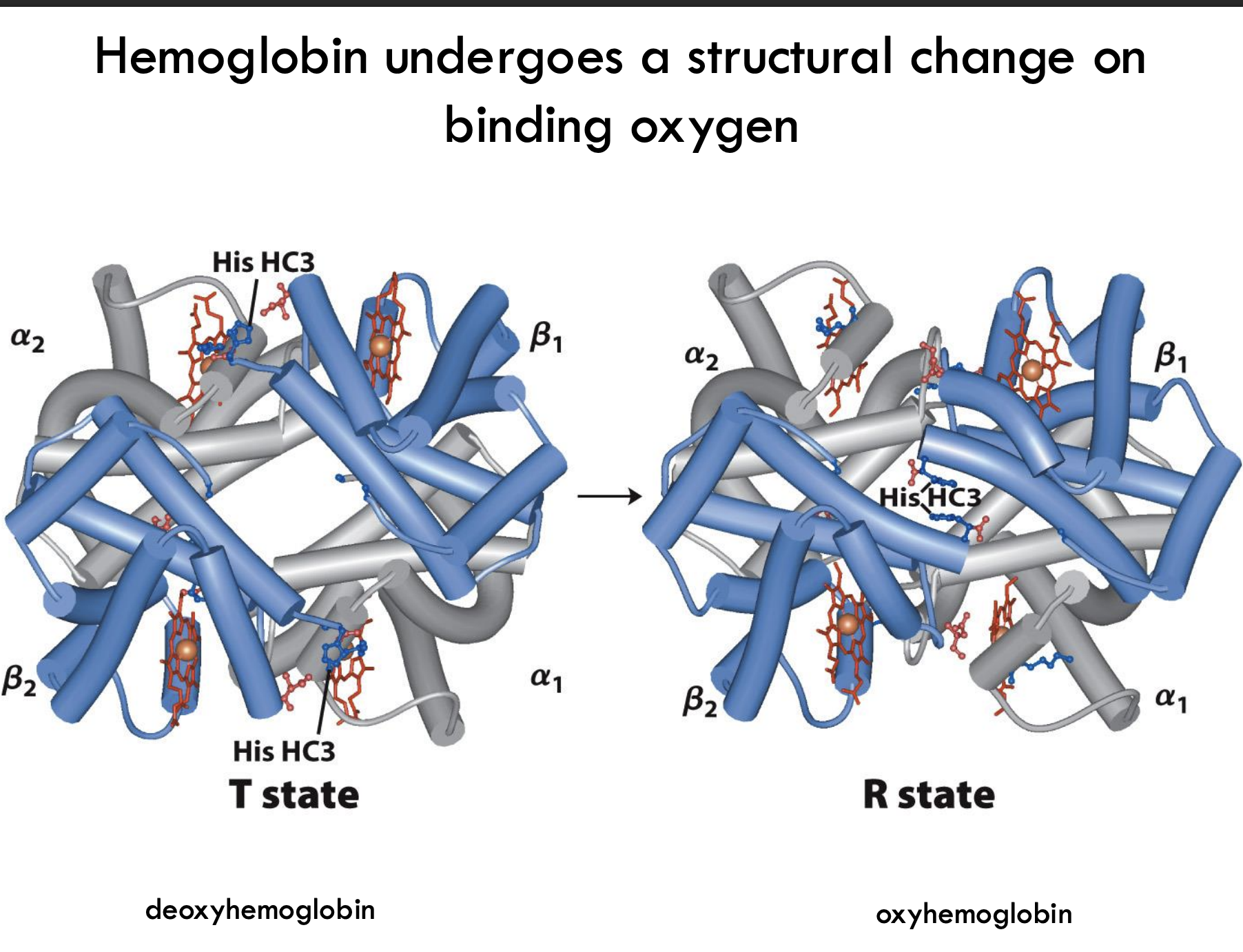
What structural change does hemoglobin undergo when binding oxygen?
Hemoglobin shifts from the T (tense) state (low oxygen affinity) to the R (relaxed) state (high oxygen affinity) when oxygen binds, causing a rearrangement of subunits and a tighter structure.
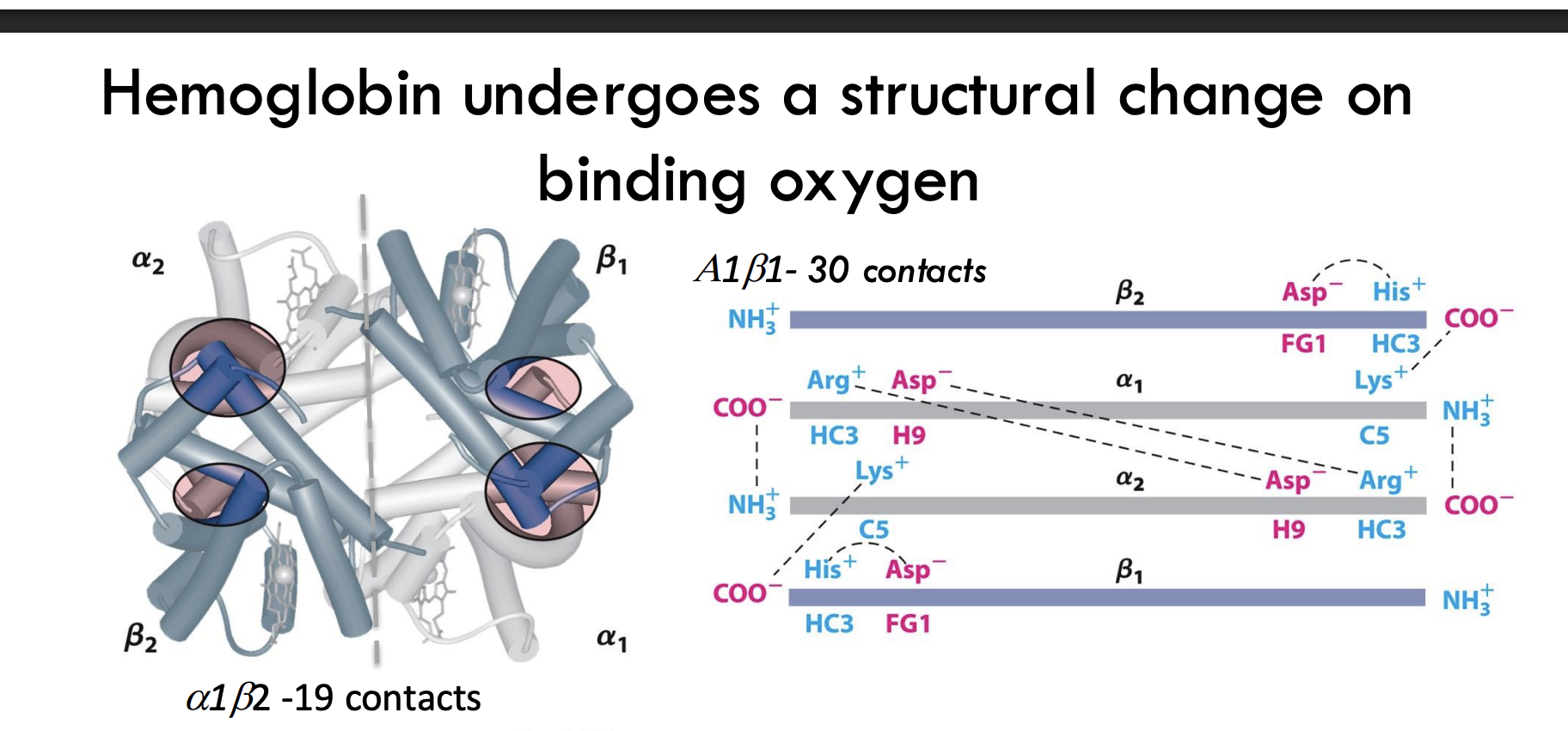
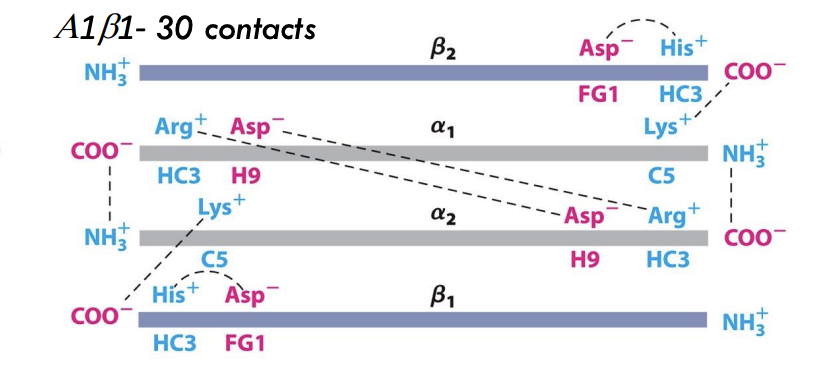
What happens to hemoglobin’s subunit contacts during oxygen binding?
Oxygen binding increases α₁β₁ and α₂β₂ subunit contacts (30 contacts) and reduces α₁β₂ and α₂β₁ contacts (19 contacts), stabilizing the R (relaxed) state.
What happens to hemoglobin if it denatures?
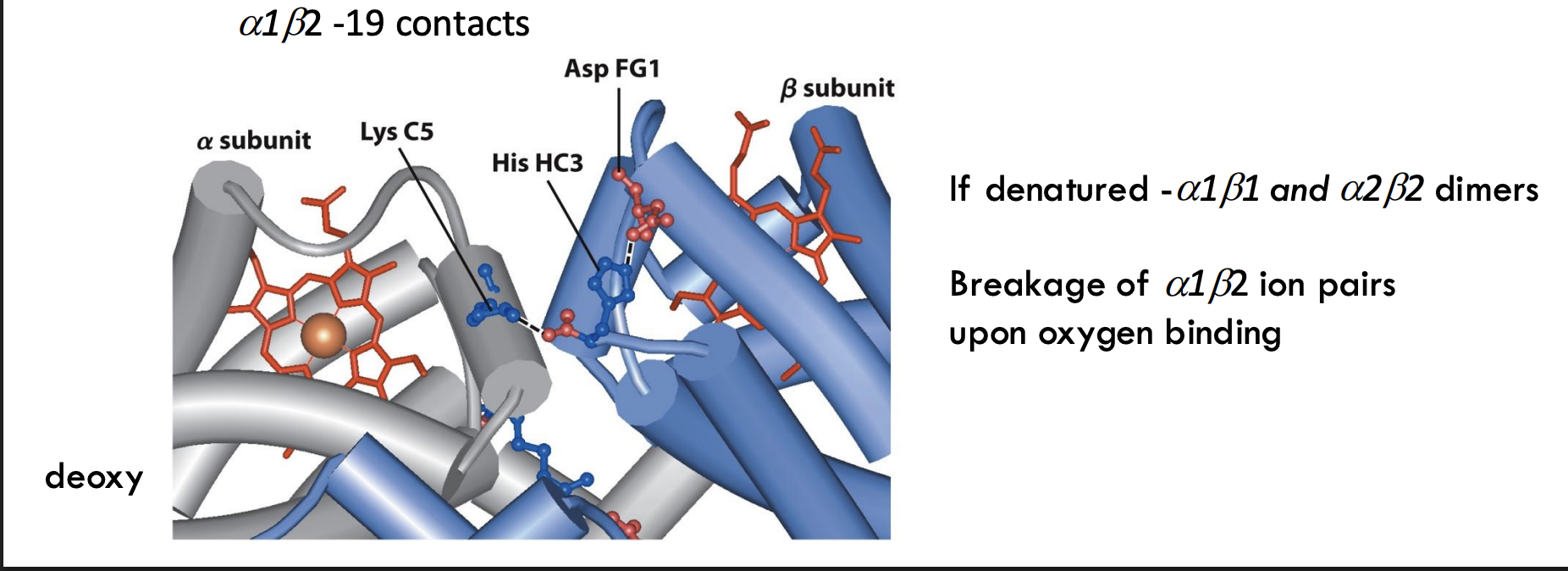
It separates into α₁β₁ and α₂β₂ dimers, meaning the two sets of subunits stay paired.
What structural event happens to ion pairs in hemoglobin upon oxygen binding?
Ion pairs between α₁ and β₂ subunits break when oxygen binds, triggering the T → R transition (tense to relaxed state).
What is the T state of hemoglobin?
The T (tense) state has more interactions, is more stable, and has a lower affinity for O₂.
What is the R state of hemoglobin?
The R (relaxed) state has fewer interactions, is more flexible, and has a higher affinity for O₂
.
What happens when oxygen binds to hemoglobin?
O₂ binding triggers a T → R conformational change, making hemoglobin more flexible and better at binding oxygen.
What structural change helps hemoglobin switch from T state to R state?
Breaking ion pairs between the α₁-β₂ interface helps shift hemoglobin to the R state after oxygen binds.
What is the MWC (concerted) model of cooperative binding?
Also called the Monod-Wyman-Changeux model
Enzyme exists in either T (tense) or R (relaxed) state
All subunits switch states together
Binding increases the chance the entire enzyme shifts to the R state (higher affinity)
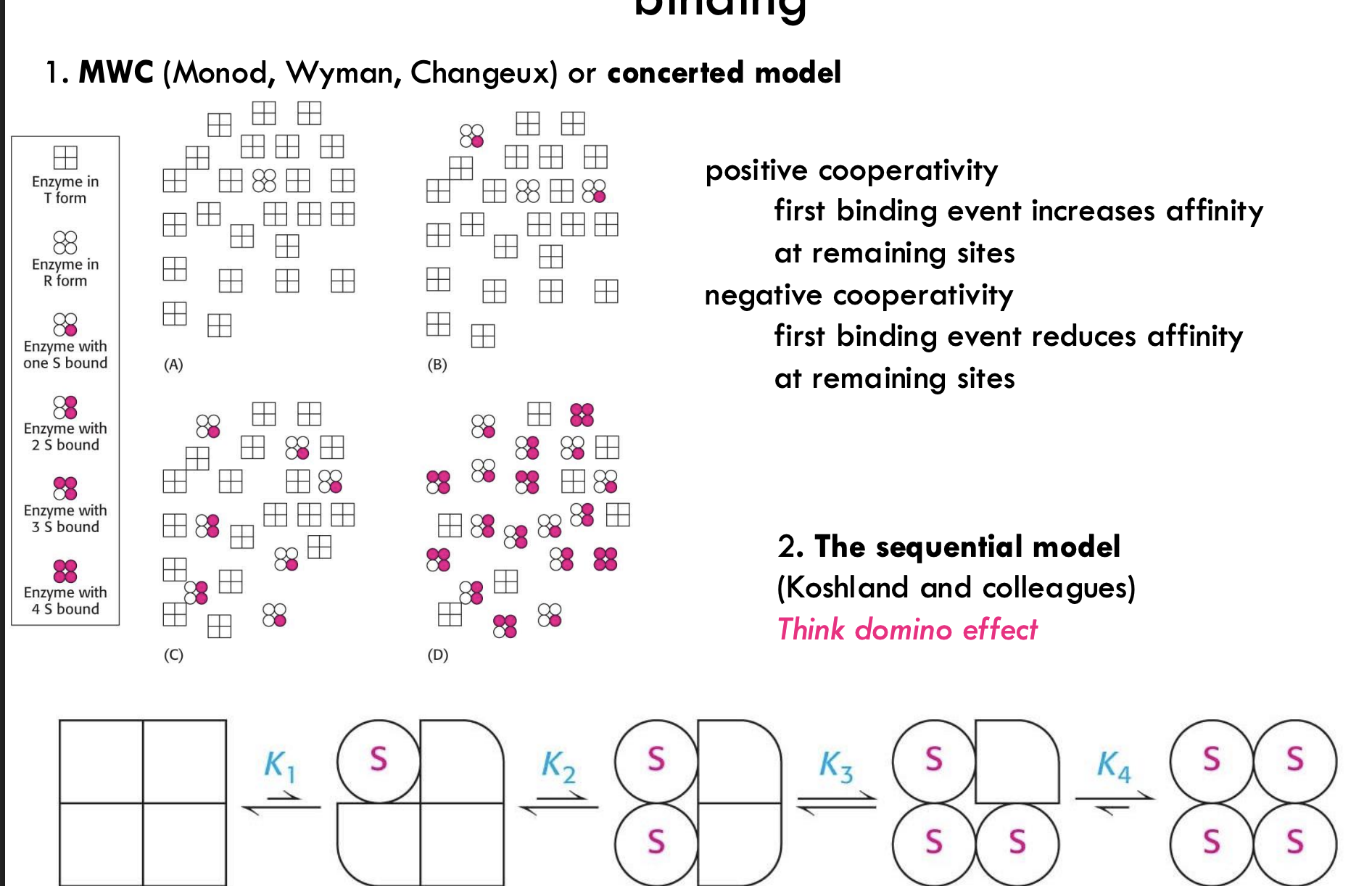
What is the sequential model of cooperative binding?
Developed by Koshland and colleagues
Subunits change one at a time as substrate binds
Think domino effect
Each binding event alters the conformation of the next site
What is positive and negative cooperativity?
Positive cooperativity: First binding makes it easier for the next
Negative cooperativity: First binding makes it harder for the next
What is the primary function of myoglobin versus hemoglobin?
Myoglobin: Oxygen storage in muscle cells
Hemoglobin: Oxygen transport from lungs to tissues
How does the oxygen binding curve of myoglobin compare to that of hemoglobin?
Myoglobin: Hyperbolic curve → binds O₂ tightly
Hemoglobin: Sigmoidal curve → shows cooperative binding, allowing efficient O₂ loading and unloading
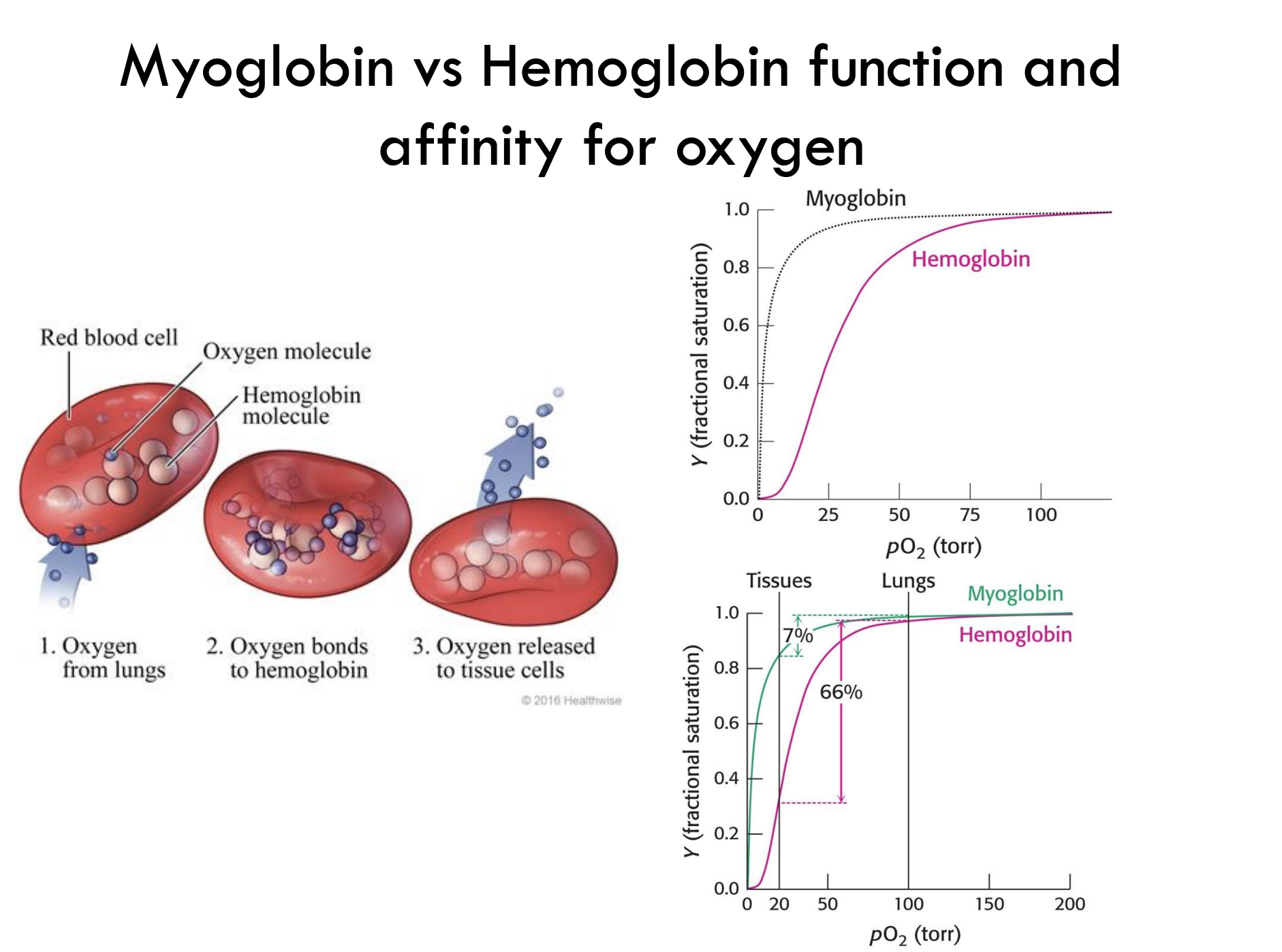
How much oxygen does hemoglobin release from lungs to tissues?
Hemoglobin releases about 66% of its bound oxygen when moving from lungs (high pO₂) to tissues (low pO₂).