Biological Molecules
1/46
Earn XP
Description and Tags
unit1 1
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
47 Terms
define a monomer and give examples
smaller units from which larger molecules are made from
→ amino acids/ monosaccharides/ nucleotides
define polymer
molecules made from a large number of monomers joined together
→ polysaccharides/ polynucleotide/ polypeptine
describe a condensation reaction
Involves the joining of two molecules (monomers) by a chemical bond
involves the elimination of a water molecule
a condensation reaction between two monosaccharides form a glycosidic bond
describe a hydrolysis reaction
hydrolysis reactions break the chemical bond (covalent) between two molecules
involves the use of water
define monosaccharaide and give examples
Monomers from which larger carbohydrates are made from
General formula (CH2O) n
→ glucose/ fructose/galactose
draw the alpha glucose and beta glucose molecules
glucose has two isomers:
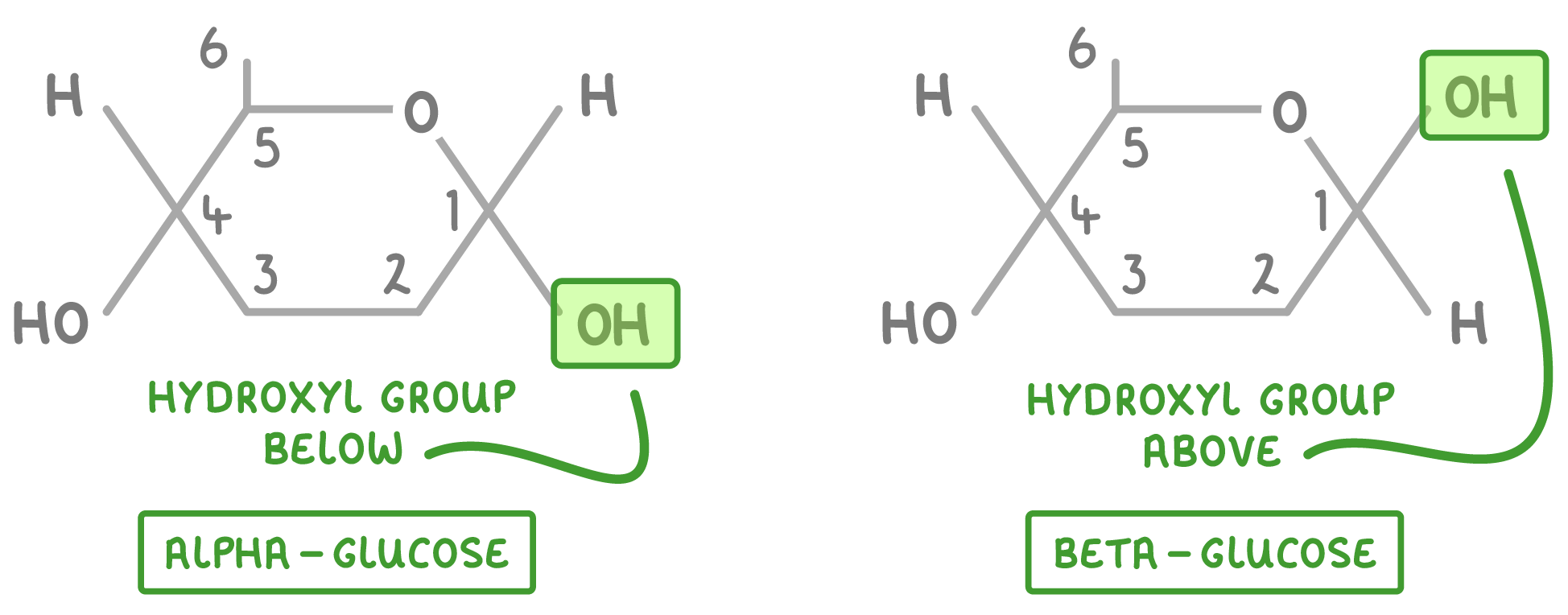
define an isomer
molecules that have the same molecular formula but a different structural formula
how are the disaccharides formed
Condensation of two monosaccharides joined together by a glycosidic bond (C12H22O11 )
glucose+ glucose → maltose + water
glucose +fructose →sucrose + water
glucose+ galactose→ lactose+ water
what are polysaccharides?
formed by the condensation of many glucose units
→ Starch/ Glycogen/ Cellulose
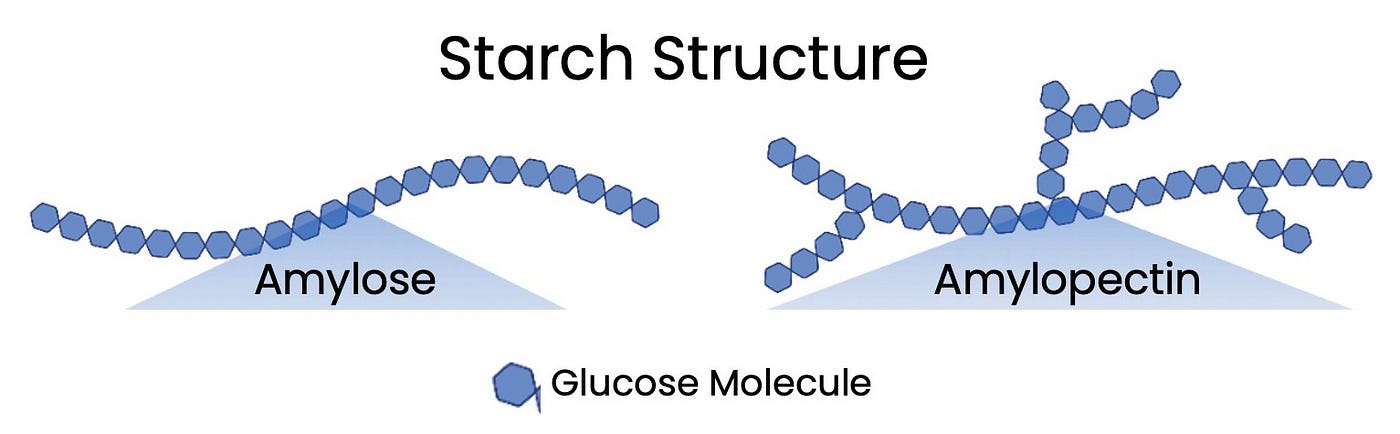
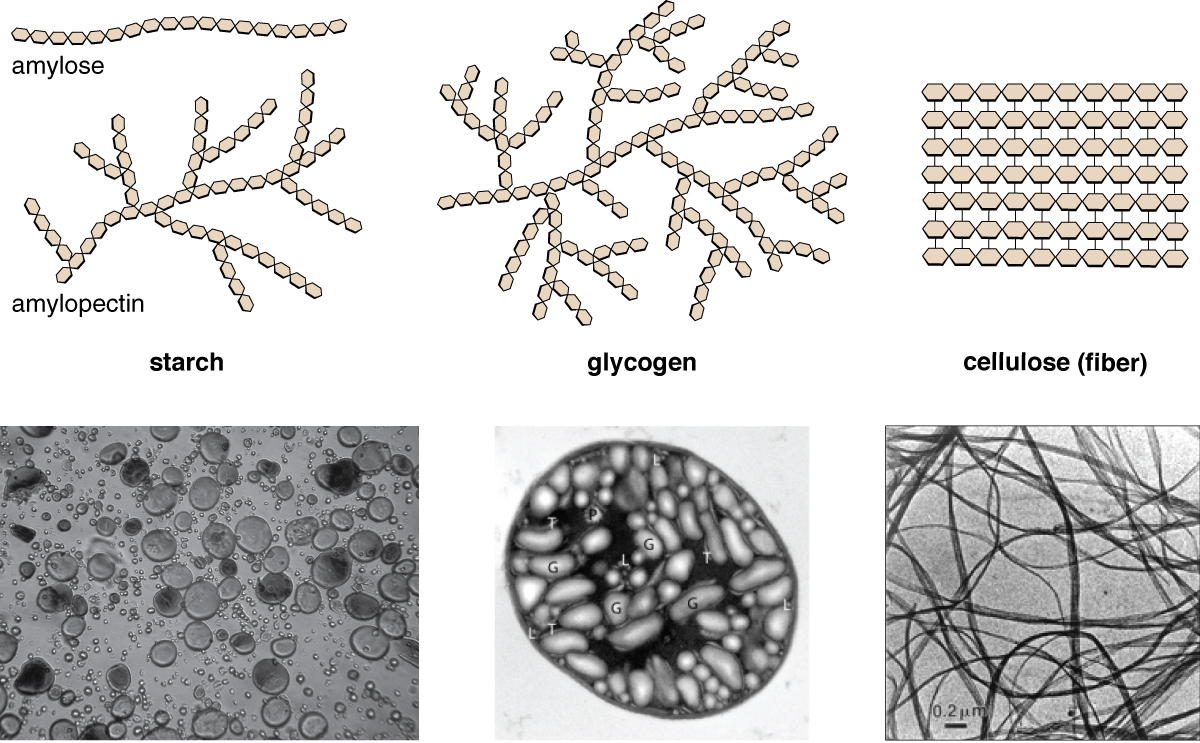
explain how the structure of starch leads to the function
made of alpha glucose monomers that form 2 polymers amylose-unbranched helix 1-4 glycosidic bondsamylopectin-branched molecule 1-4 and 1-6 glycosidic bond
coiled/ helical can compact to fit many glucose
branched structure increases SA for rapid hydrolysis back to glucose
glucose easily accessible by enzymes to break down for respiration
insoluble so osmotically inactive
Large so cannot cross cell-surface membrane
explain how the structure of cellulose leads to the function
polymer forms long straight chains of beta glucose monomers that are only joined by 1-4 glycosidic bond
chains are held in parallel by many hydrogen bonds to form a fibril structure
many hydrogen bonds provide collective strength
insoluble (wont affect water potential)
can resist osmotic pressure
bond is difficult to break
resists actions of enzymes
describe the test for non-reducing sugars
do benedicts test + solution stays blue/negative
Add the sample in a test tube, add dilute HCL and boil for a few minutes to hydrolyse the glycosidic bond
Add sodium hydrogen carbonate powder to neutralise the solution
Add benedicts reagent, heat the mixture gently in a waterbath
how are triglycerides formed why aren’t they polymers
Condensation reaction between one molecule of glycerol and three molecules of fatty acid forms ester bonds
no repeating monomers
what’s the difference between saturated and unsaturated fatty acids
saturated-hydrocarbon chain has only single bonds
unsaturated-consists of at least one double bond between carbons
what are the properties of triglycerides
high ratio of carbon-hydrogen bonds→ releases more energy
low mass-energy ratio→ good storage molecule as doesn’t increase mass
large non-polar molecules→ insoluble in water+ waterproof
high ratio of hydrogen-oxygen bonds→ releases water when metabolised (source of water for dessert animals)
Compare and contrast the structure and properties of triglycerides and phospholipids
Both contain ester bonds between glycerol and fatty acid
Both contain glycerol
Fatty acids o both may be saturated or unsaturated
Both are insoluble in water
Triglyceride has 3 fatty acids and phospholipids have 2
Triglycerides are hydrophobic and phospholipids have hydrophilic and hydrophobic region
Phospholipids form a monolayer in water but triglycerides don’t
describe the emulsion test
dissolve in alcohol(ethanol)
add distilled water and shake
white milky emulsion appears if lipid is present
How are phospholipids formed and describe the structure
two fatty acids bond to glycerol molecule via two condensation reactions forming two ester bonds
a phosphate molecule replaces the third fatty acid chain is covalently bonded to glycerol
hydrophilic head-attracts water
hydrophobic tail-repel water
→forms the phospholipid bilayer structure
what are proteins and describe the amino acid structure
diverse group of large and complex molecules ,made from long chains of amino acids
How are polypeptides and dipeptides formed
condensation reaction of many amino acids joined together by peptide bonds and this involves the formation of water
Condensation of two amino acids Joined by a peptide bond
H from the amine group
OH-from carboxyl group
describe the biuret test
make the solution alkaline by adding a few drops of sodium hydroxide solution
add copper(II)sulphate→ copper bonds to peptide bonds
solution turns purple if protein is present
Give a description of each structural level of proteins
primary: Sequence of amino acids and determines the ultimate shape and function
secondary: Chain is twisted to form alpha helix or beta pleated sheets→ hydrogen in NH is slightly positive and oxygen in C=O is slightly negative so hydrogen bonds are formed
Tertiary structure: further folding into 3D shape held by-hydrogen bonds/ionic bonds(carboxyl+ amine group) or disulfide bridges(sulfur in R group)
Quaternary structure: made of two or more polypeptide chains
what is an enzyme
globular proteins which are biological catalysts to specific biological reactions
describe the induced fit model
The shape of the active site of an enzyme changes when the substrate binds to it
the enzyme becomes complementary
How does the active site of an enzyme cause a high rate of reaction
In the Enzyme-Substrate complex there’s a change in shape of the active site
Causes pressure to be placed on the bonds within substrate→ they begin to break
reduces activation energy
describe the effect of substrate concentration on rate of reaction
At low substrate conc= low rate of reaction, some active sites are free but limited chance of successful collision
At intermediate substrate conc= high rate, more substrate molecules so high chance of successful collision
High substrate conc= addition of more substrates has no effect as all active sites are saturated
describe the effect of enzyme concentration on RoR
low enzyme conc- too few enzymes, not all substrates have active site so half possible RoR
intermediate enzyme concentration- RoR increases as all active site full ,more substrate can occupy active site→ form products
Addition of further enzymes have no effect on RoR as there are more enzyme than substrate
describe the effect of temperature on RoR
increase in kinetic energy, more successful collisions
past optimum-higher kinetic energy break bonds in tertiary structure
→weaker hydrogen/ionic bonds
shape of active site denatures, enzyme loses catalytic ability so no more ES complex
describe how PH effects RoR
at optimum PH many collisions between enzymes and substrate→ high RoR
change in PH means more or less ions in a solution, this affects the bonds in the tertiary structure
active site becomes denatured, no successful collisions
how do competitive inhibitors lower ROR
inhibitor has a similar shape to the substrate
inhibitor is able to bind to active site
fewer ES complexes formed
→ lower ROR
how do non-competitive inhibitors lower ROR
inhibitor binds to a seperate site on the enzyme
changing the shape of active site
substrate can no longer bind so cannot form ES complex
how do inhibtors affect rate of reaction with increasing substrate concentration
with competitive inhibitors the reaction reaches max rate but only at higher substrate concentration
higher chance of collisions forming ES complex
with non-competitive inhibitor reaction cannot reach max rate
How is DNA found in eukaryotic cells + prokaryotic cells
packaged as chromosomes in the nucleus
DNA associated with histones proteins→chromatins→chromosomes
Most RNA in cytoplasm particularly ribosomes
Structure of DNA Nucleotide
phosphate group
Deoxyribose sugar, Pentose
Nitrogenous bases: adenine, thymine, guanine, cytosine
3 components join together to form a mono nucleotide via condensation (using ATP)
Structure or RNA nucleotide
phosphate group
Ribose sugar
Nitrogenous bases: Adenine, Uracil, guanine, cytosine
Single stranded+ shorter than DNA
3 types of RNA
mRNA→ messenger, carries DNA code from nucleus to ribosome
tRNA→Transfer, carries amino acids across the cytoplasm
rRNA→ ribosomal, makes up the ribosome
How is the double helix structure of DNA formed
the nucleotides join together via a condensation reaction between the phosphate group of one nucleotide and the sugar of another →phosphodiester bond
Two DNA polynucleotide strands are joined by hydrogen bonds between the bases
Two hydrogen bonds between A-T and three hydrogen bonds between C-G
Describe the process of semi-conservative replication
DNA Helicase breaks hydrogen bonds between bases
Each separated strand acts as a template
Free DNA nucleotides align by complementary base pairing
DNA polymerase joins adjacent DNA nucleotides in a condensation reaction using ATP forming phosphodiester bonds
Each new DNA strands are formed each with one original strand and one new strand
Chargaffs rules of base pairing
Number of guanine units= number of cytosine units
Number of adenine units= number of thymine units
How is DNA adapted for its function
sugar-phosphate backbone→very stable
Weak hydrogen bonding between bases→ can easily separate for DNA replication
Many hydrogen bonding→ stable structure
Extremely large molecule→ can store a lot of genetic information
Double stranded→ strand can act as templates for semiconservative replication + allows accurate complementary base pairing
Coiled→so can compact, good storage molecule
Base sequence→ Allows for amino acid coding
Structure of ATP
3 phosphate molecules
Pentose sugar molecule, ribose
Adenine nitrogen containing compound
Equation for ATP Synthesis
ADP + Pi→ ATP + H20 catalysed by ATP synthase
H20 + ATP→ ADP+ Pi catalysed by ATP hydrolase
How is energy stored in ATP
Bonds between phosphate groups are unstable
Lower Ea
Can easily be broken down
Release a small amount of energy
Advantages of ATP
releases energy in small manageable amounts
Can be reformed again
One single chemical reaction→ energy can be released quicker
Can phosphorylate other molecules→ making them more reactive
Does not move out of the cell
What are the properties of water
metabolite→Allows metabolic reactions to occur
Polar solvent→ allows metabolic processes to occur faster
High SHC→ resists temperature changes
High latent heat of vaporisation→ when evaporated it has a cooling effect
Transparent→ allows photosynthesis of aquatic plants to occur
Adhesive+ cohesive → allows water to be pulled up the xylem
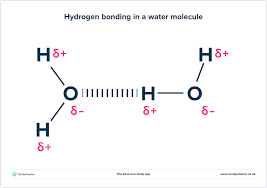
Draw the interaction of water molecules
Describe the roles of iron ions, sodium ions and phosphate ions in cells
Iron: haemoglobin associates with oxygen
Sodium:
Co-transport of glucose/ amino acids into cells
Na+ moves out by active transport
Creates sodium concentration gradient
Affects osmosis/ water potential
Phosphate ions:
Affects osmosis/ water potential
Joins nucleotides in phosphodiester bonds
Used in ATP
Phosphorylates other compounds
Hydrophilic part of phospholipid bilayer