Gene Expression Study Guide (BIO 161)
1/140
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
141 Terms
Genes
Units of genetic information (DNA) that carry instructions for building polypeptides (proteins) or functional RNA molecules along with regulatory sequences
Gene Expression
Process of converting archived information into molecules that actually do things
The Central Dogma of Molecular Biology
Summary of the flow of information in cells
Transfer RNA (tRNA)
"Interpreter" molecule; transfers amino acids to the ribosomes
Ribosomal RNA (rRNA)
Component of RNA
Information flow is always in one direction
False. In some cases, information flows from RNA back to DNA
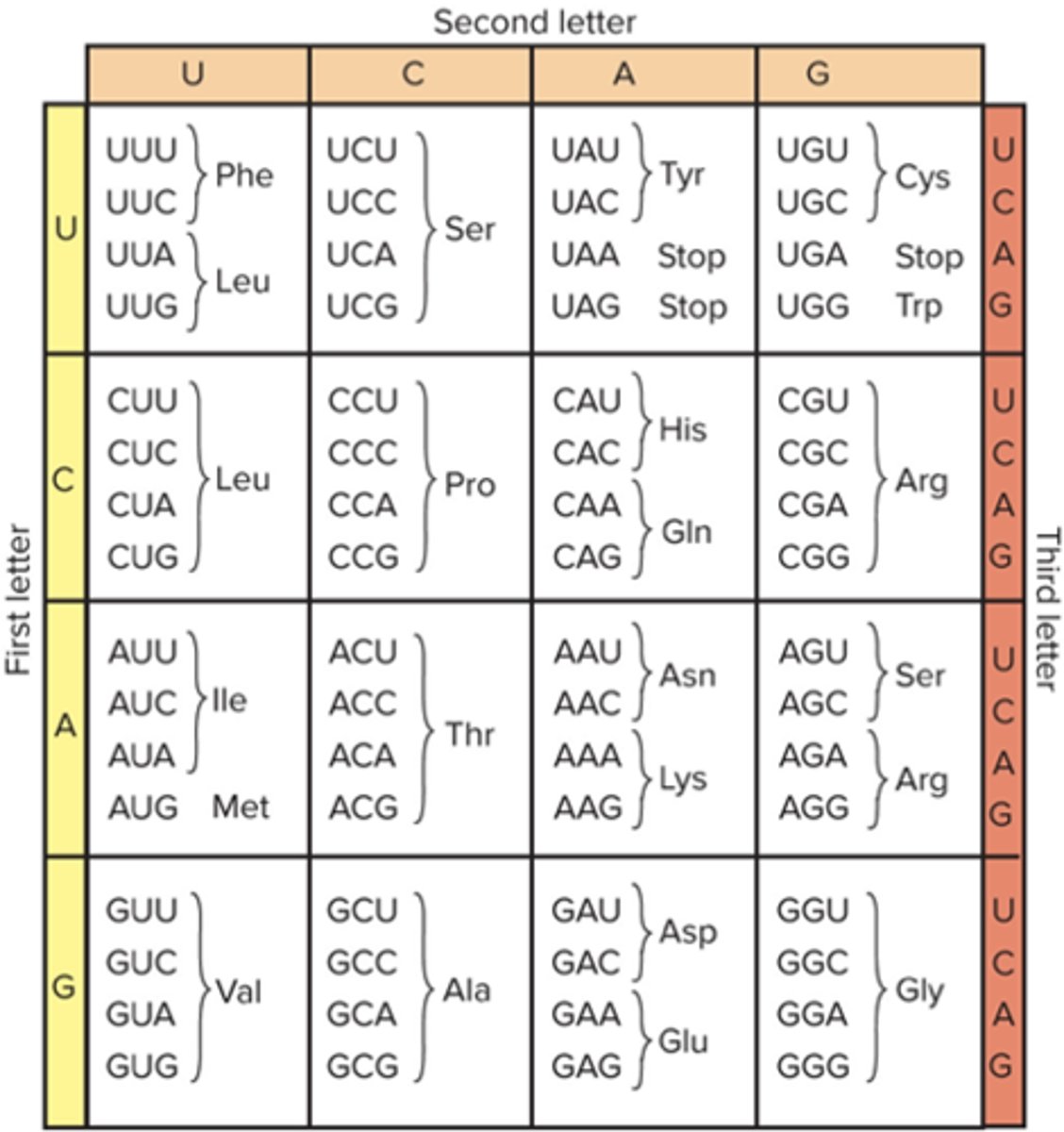
How many nucleotide bases are there?
4 (A,T,G,C)
How many amino acids do the nucleotide bases specify?
20
Triplet Code
Each amino acid is code for by a group of three bases
Codon
Group of three bases that specifies a particular amino acid
Start Codon
Identifies the site at which protein synthesis should start; codes for methionine (AUG)
Stop Codons
Signify that protein synthesis is complete (UAA, UAG, UGA0
What does it mean when genetic code is redundant?
More than one triplet may specify the sam amino acid
What does it mean when genetic code is unambiguous?
Each codon has only one meaning
What does it mean when genetic code is conservative?
First two bases of codons that specify the same amino acid are usually identical
What does it mean when genetic code is universal?
The same genie code is used by all living things
Translate this template strand:
3' - CGTACCAGTTCGCATCGATTT - 5'
5' - GCAUGGUCAAGCGUAGCAAA - 3'
Mutation
Any permanent change in an organism's DNA
Point Mutation
Replacement of one nucleotide with another (Silent, Missense, Nonsense)
Silent Mutation
Does not alter amino acid sequence
Missense (Substitution) Mutation
Changes one amino acid to another
Nonsense Mutation
Changes codon for an amino acid to STOP codon (polypeptide chain to short = non functional protein)
Frameshift (Insertion or Deletion) Mutation
Alter reading frame (group of codons) of mRNA triplets resulting in non-functional protein
Chromosomes
DNA and proteins (histones) packed together
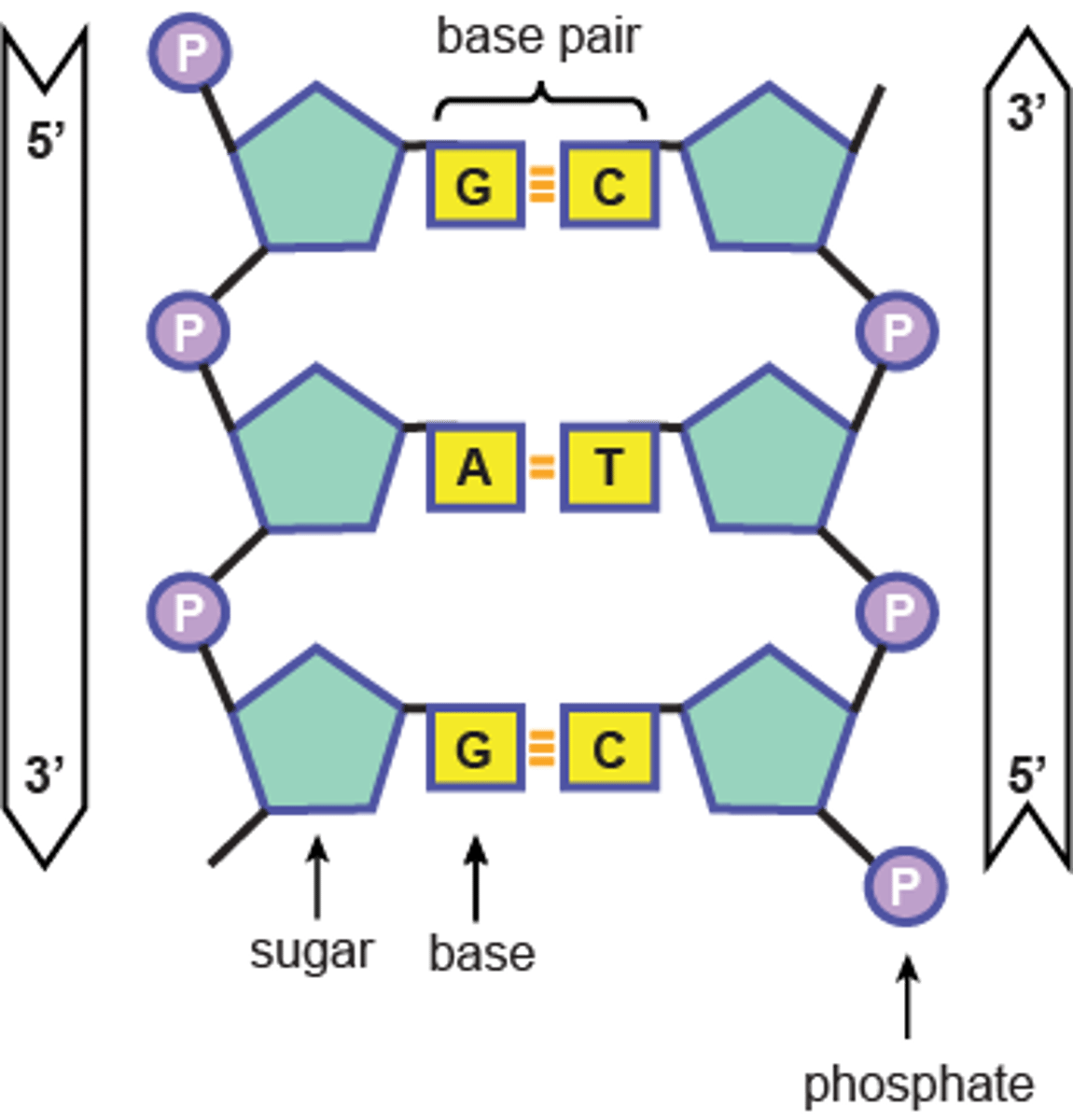
Double Helix
Two polynucleotide strands, twist about one another with a sugar-phosphate backbone
Complementary Base Pairing
Bases hydrogen bond with one another (A pairs with T, G pairs with C)
DNA Replication
Making a copy of the DNA in a cell
How does DNA replicate?
DNA unwinds and unzips where a old strand serves as a template and complementary nucleotides are added to form a new strand

Daughter Strands
Identical to parent helix containing one old strand and one new strand
What is the complementary strand for the template strand?
5' - ATCCTCG - 3'
3' - TAGGAGC - 5'
DNA Polymerase
Catalyzes the addition of nucleotides to existing 3'-OH groups
Does DNA polymerase require a primer?
Yes
Primers
Short RNA starters are synthesized by primase (an RNA polymerase)

Prokaryotes
Single origin of replication
Eukaryotes
Multiple points of orgin
What direction does DNA synthesis occur?
5' to 3' direction
Helicase
Catalyzes the breaking of hydrogen bonds between base pairs and the opening of the double helix
Topoisomerase
Breaks and rejoins the DNA double helix to relieve twisting forces caused by the opening of the helix
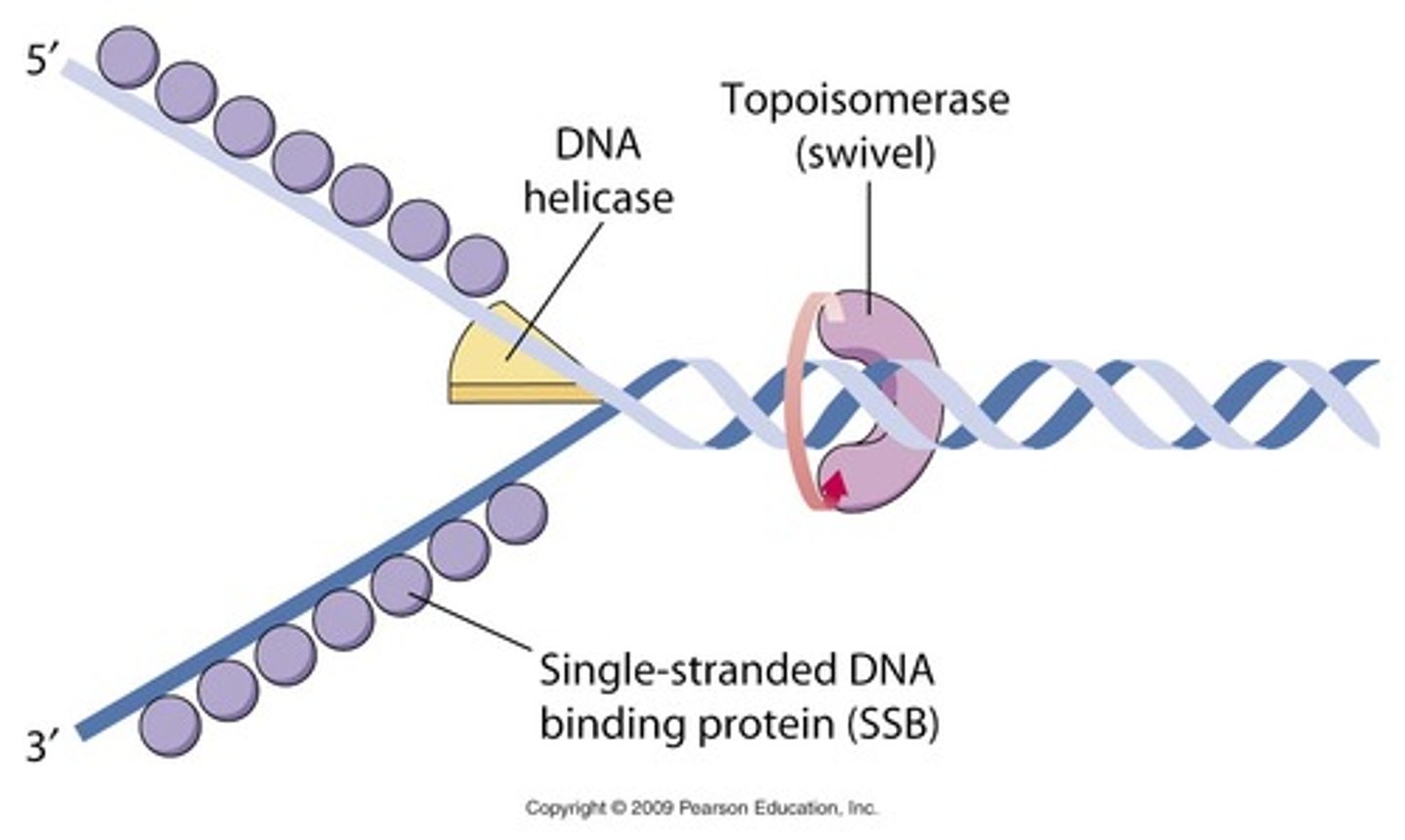
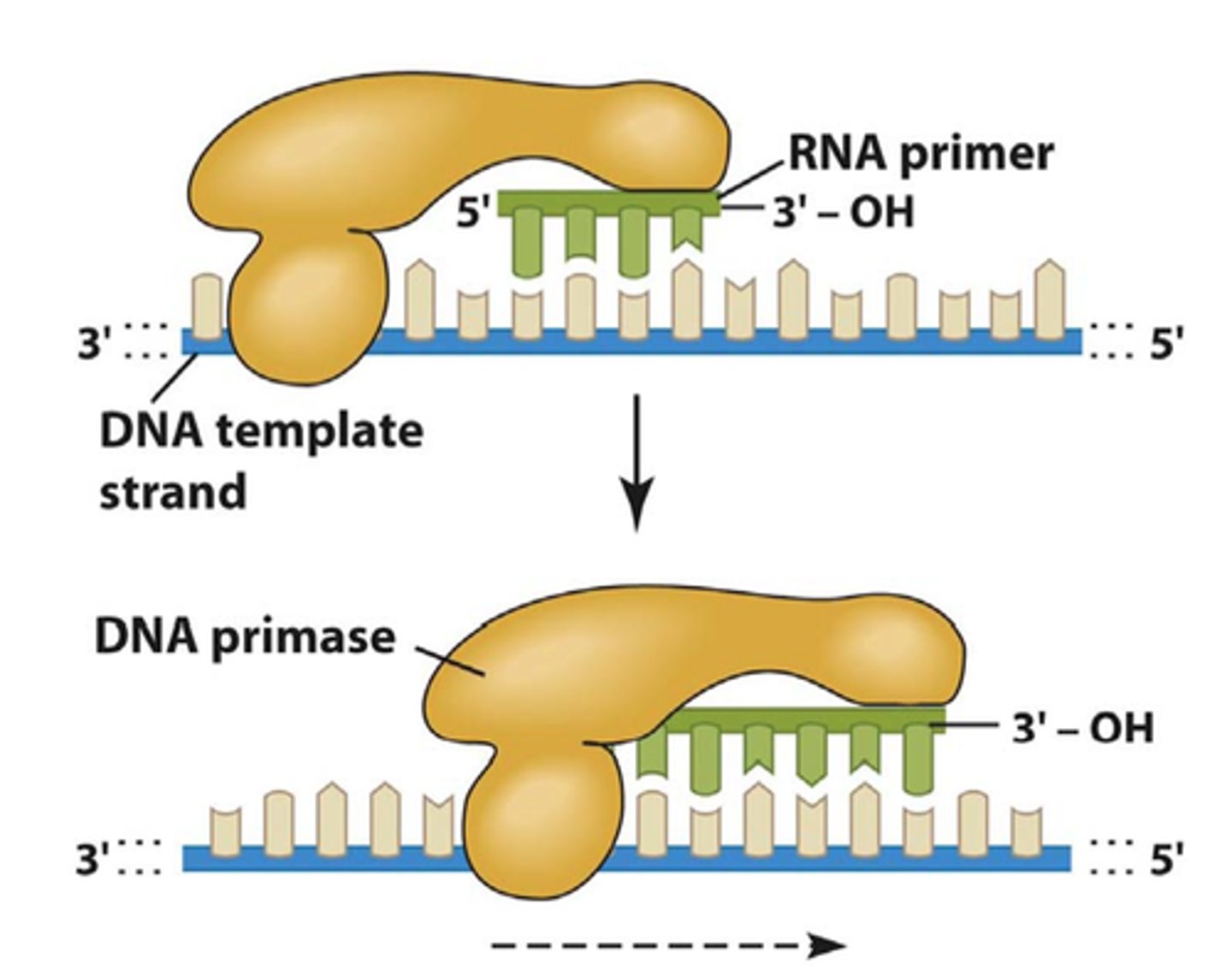
Primase
Catalyzes the synthesis of the RNA primer
DNA Polymerase III
Extends the leading strand
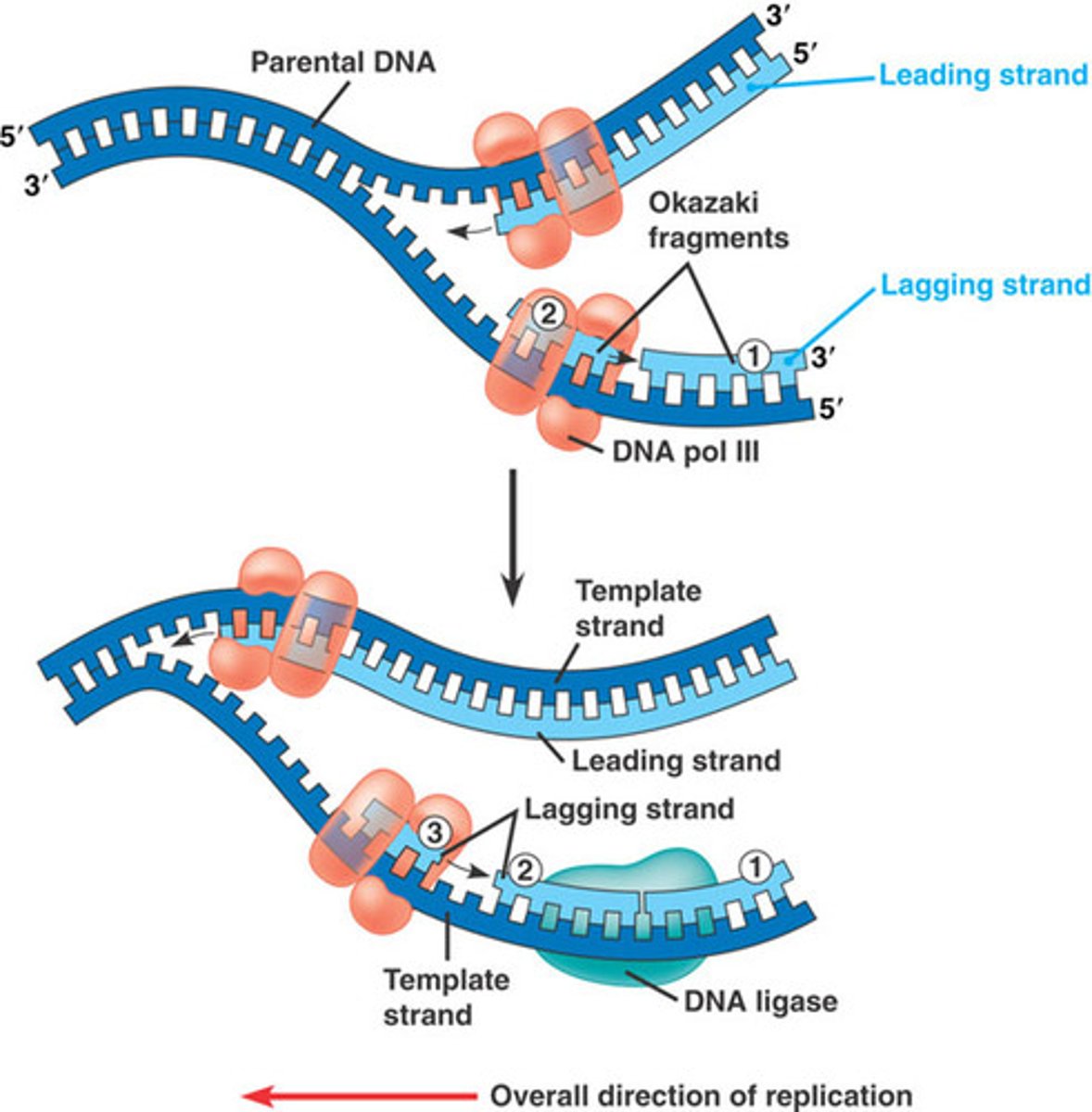
Lagging Strand Synthesis
Discontinuous; synthesize a series of 100-200 bp Okazaki fragments
DNA Polymerase I
Removes the RNA primer and replaces it with DNA
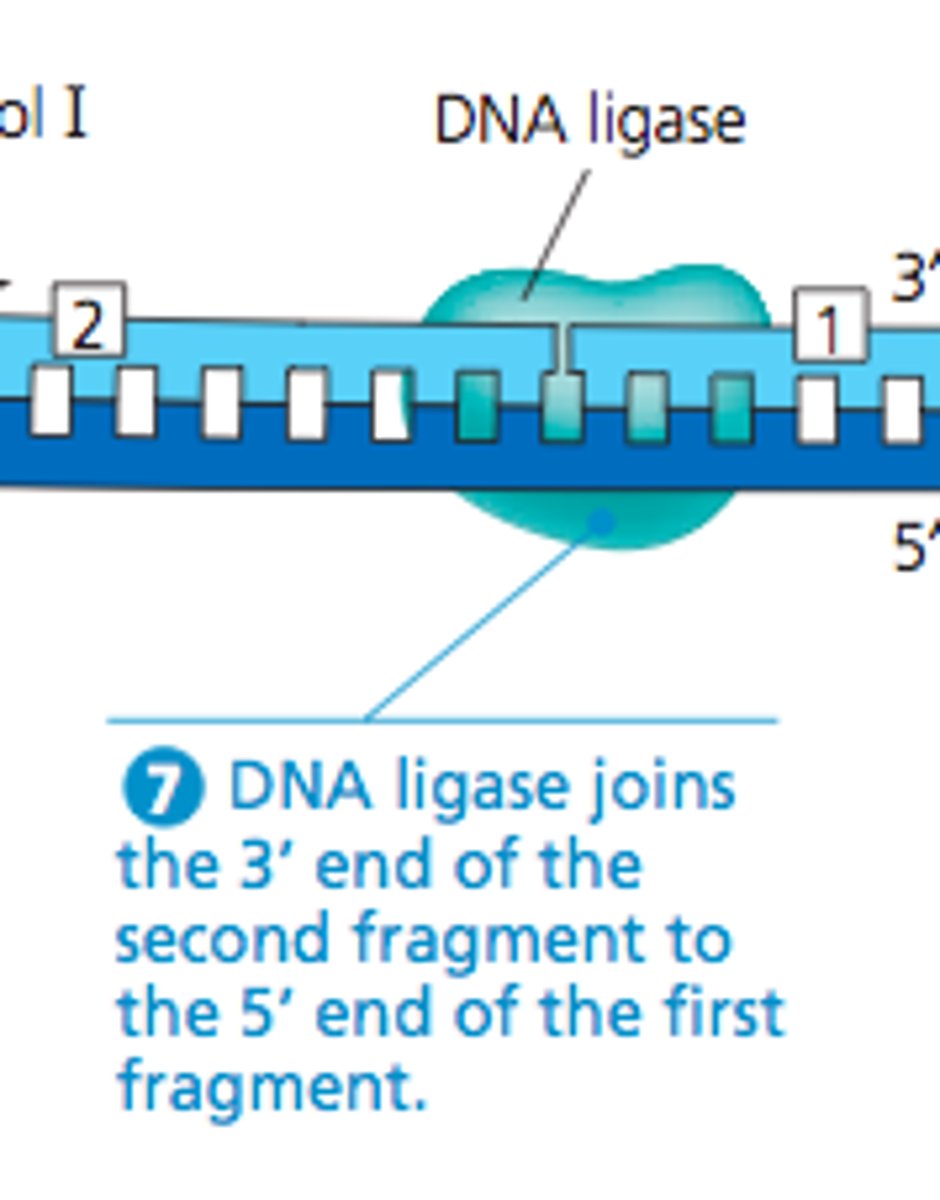
DNA Ligase
Catalyzes the joining of Okazaki fragments into a continuous strand
What are some problems that can occur with DNA Synthesis?
Chromosome shortening, mismatched bases, damages bases
Telomeres
The ends of linear chromosomes
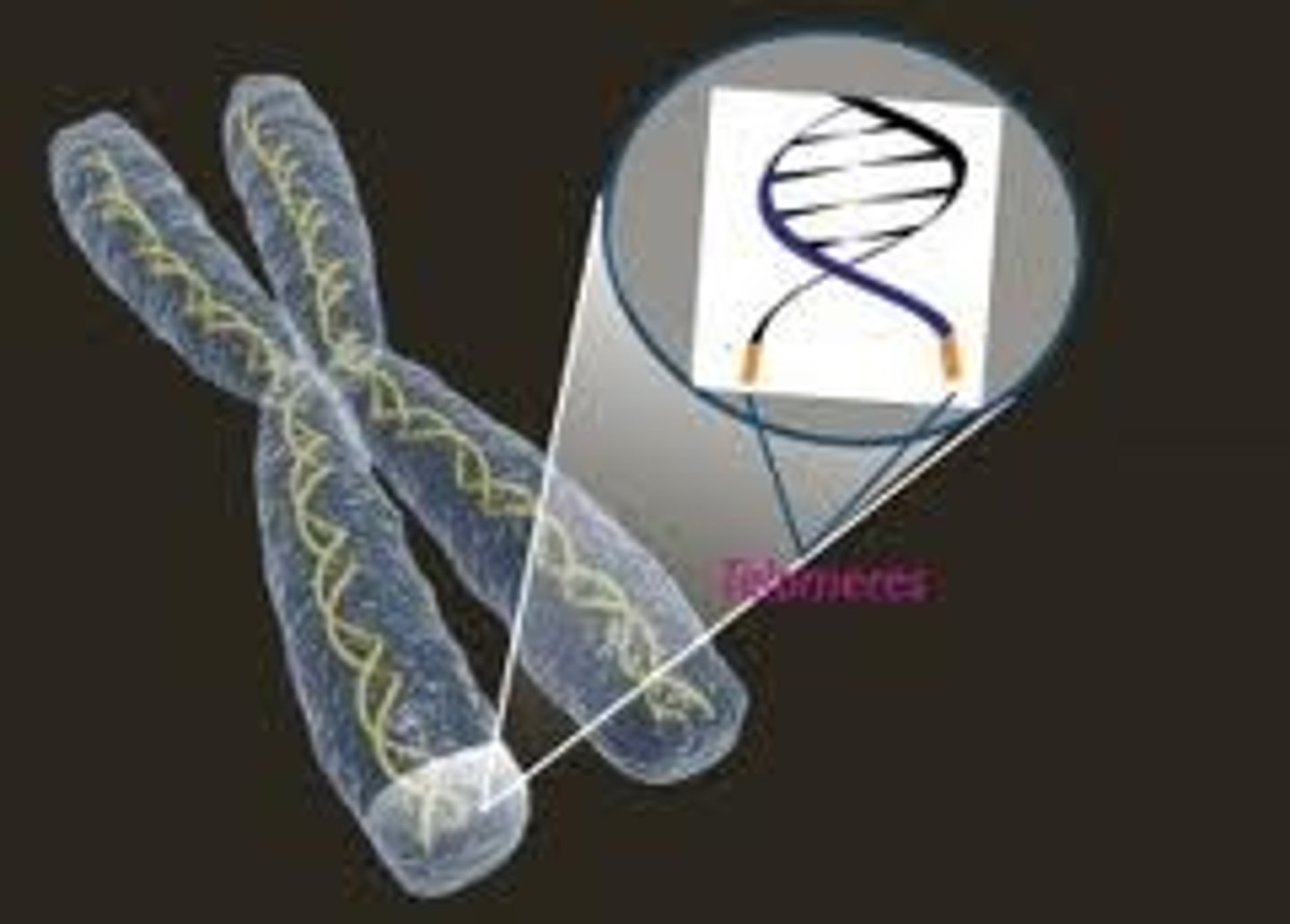
Telomerase
Replicates the ends of chromosomes
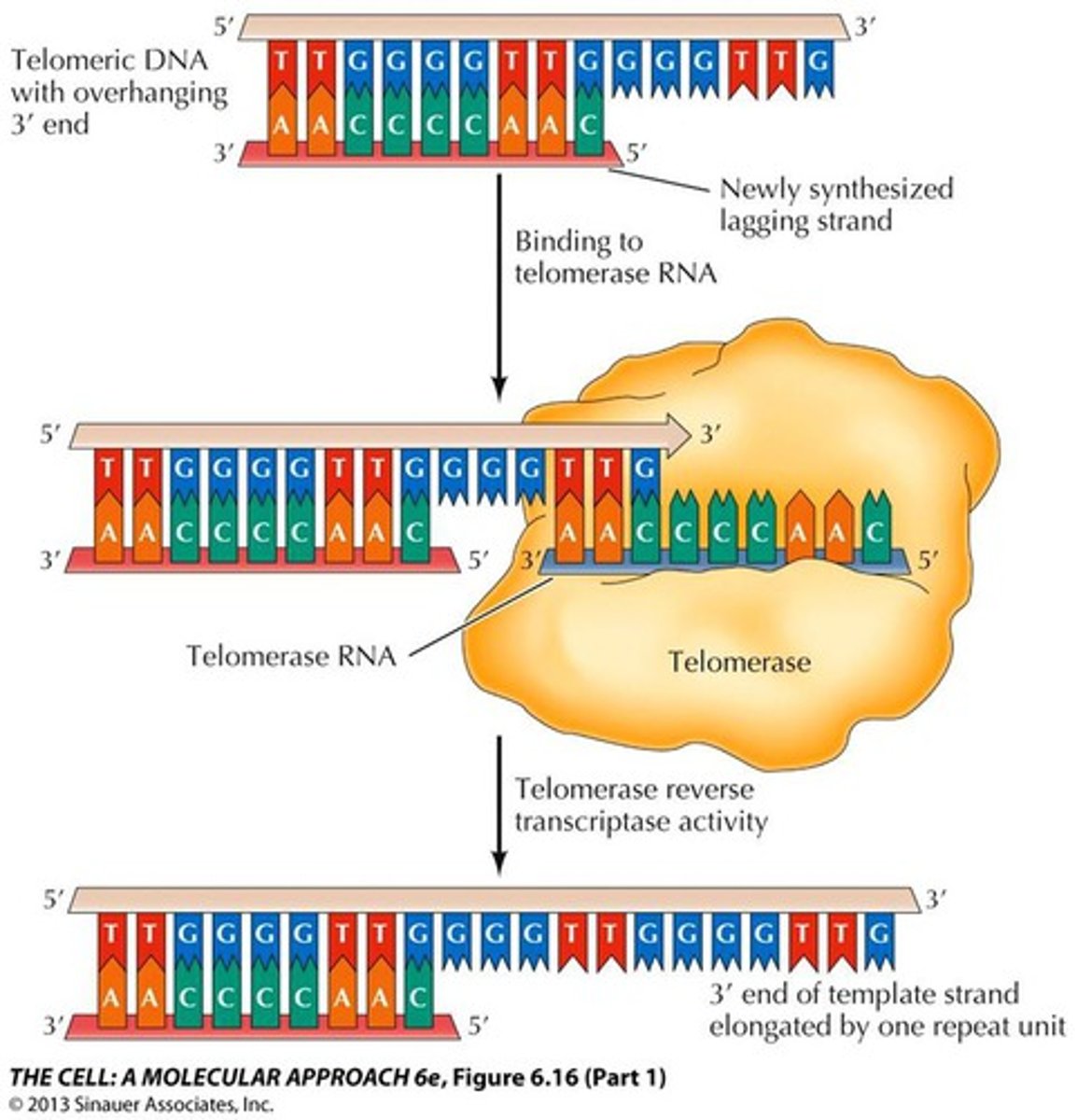
Mismatch Repair
DNA polymerase can repair mismatched bases after replication
Nucleotide Excision Repair
Removes and fixes damaged or wrong nucleotides (bases)
Transcription
Process by which messenger RNA (mRNA) is made from a DNA template
Translation
Process by which proteins and peptides are synthesized from mRNA
What direction is new RNA strand "built"?
5' to 3'
What direction does RNA polymerase travel along DNA template strand?
3' to 5'
Where does RNA polymerase begin transcribing?
At a promoter
Promoter
Part of DNA, initiation site of transcription
What are transcription factors and what do they do?
They are proteins, bind to promote region of DNA, and recruit RNA polymerase II
Transcription initiation in Prokaryotes
Sigma factors bind to promoter
Transcription initiation in Eukaryotes
Basal Transcription Factor binds to promoter, and Regulatory Transcription Factor binds to enhancer; together they recruit RNA polymerase
RNA Polymerase
Opens the helix; transcription begins (does not need a primer)
Transcription elongation
Sigma factor/transcription factor is released: RNA polymerase moves along DNA 3' -> 5', synthesizing RNA in the 5' -> 3' direction
Transcription termination in Prokaryotes
Transcription stops when RNA polymerase reaches a termination sequence (codes for RNA that forms a hairpin)
Termination in Eukaryotes
Transcription termination is triggered at poly(A) signal sequence; then a tail of hundreds of "A" is added to the mRNA
Relationship between transcription and translation in Prokaryotes
Transcription and translation are tightly coupled
Relationship between transcription and translation in Eukaryotes
Transcription and translation are separated in space and time
Which cell uses mRNA processing?
Eukaryotes
mRNA Processing
Occurs in the nucleus before mRNA is exported to the cytoplasm for translation
What happens in mRNA processing?
Addition of 5'-cap, addition of 3'-polyA tail, removal of introns
5'-cap
Composed of modified guanine nucleotide; serves as a recognition signal for translation machinery (ribosome)
3'-poly(A) tail
Composed of 100-250 adenine nucleotides; facilitates transport out of nucleus; protects mRNA message from degradation in the cytoplasm
Exons
Expressed (coding) regions
Introns
Intervening (noncoding) regions
RNA Splicing
Sp.icing is catalyzed by small nuclear RNAs and small nuclear ribonucleic proteins - snRNAs and snRNPs
What does splicing form?
A multiprotein complex called spliceosome
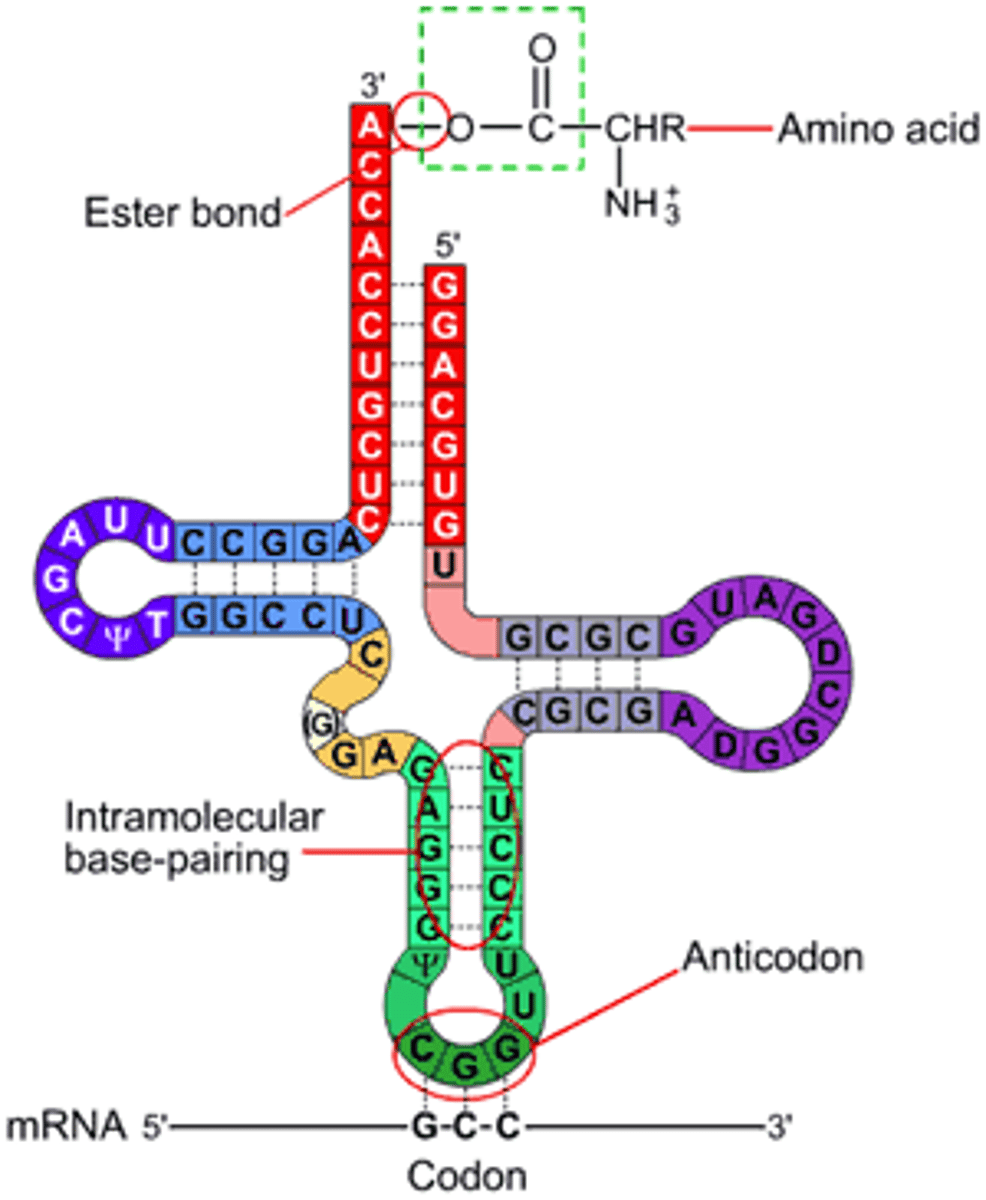
tRNA Structure
Made up of about 80 nucleotides of RNA that was transcribed from DNA
Are all tRNA's alike?
NO
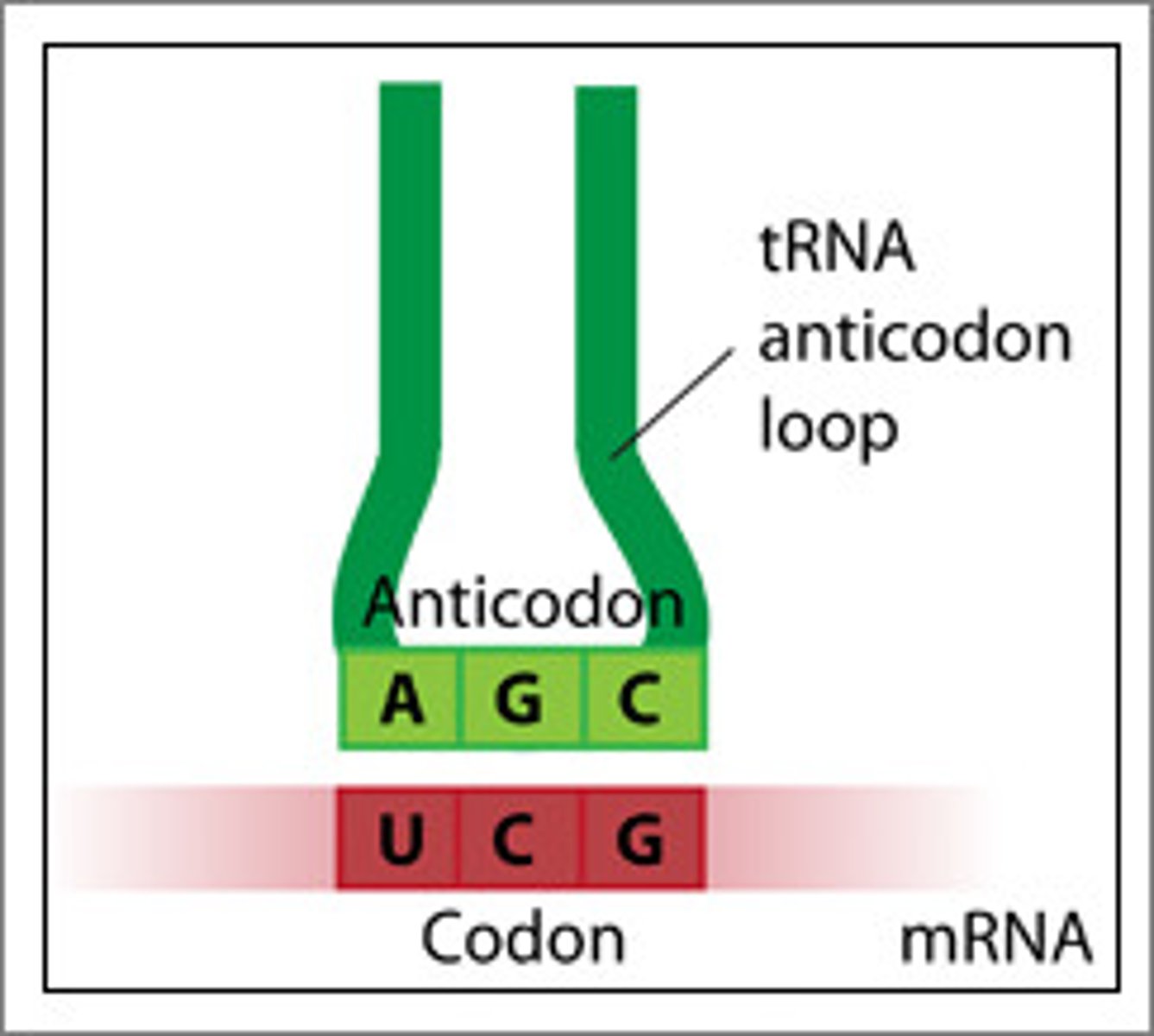
Anti-codons
Pair with codons which determines what amino acid gets attached on top of tRNA
Aminoacyl-tRNA Synthestase
Enzyme that reads the anticodon on the tRNA and then puts the correct amino acid on
Does aminoacyl-tRNA synthetase require ATP?
Yes
Are aminoacyl-tRNA synthetase specific?
Yes they are specific for each amino acid and its tRNAs
Ribosome
Site of protein synthesis; composed of ribosomal RNA (rRNA) and proteins
Small subunit (in ribosomes0
Holds mRNA in place
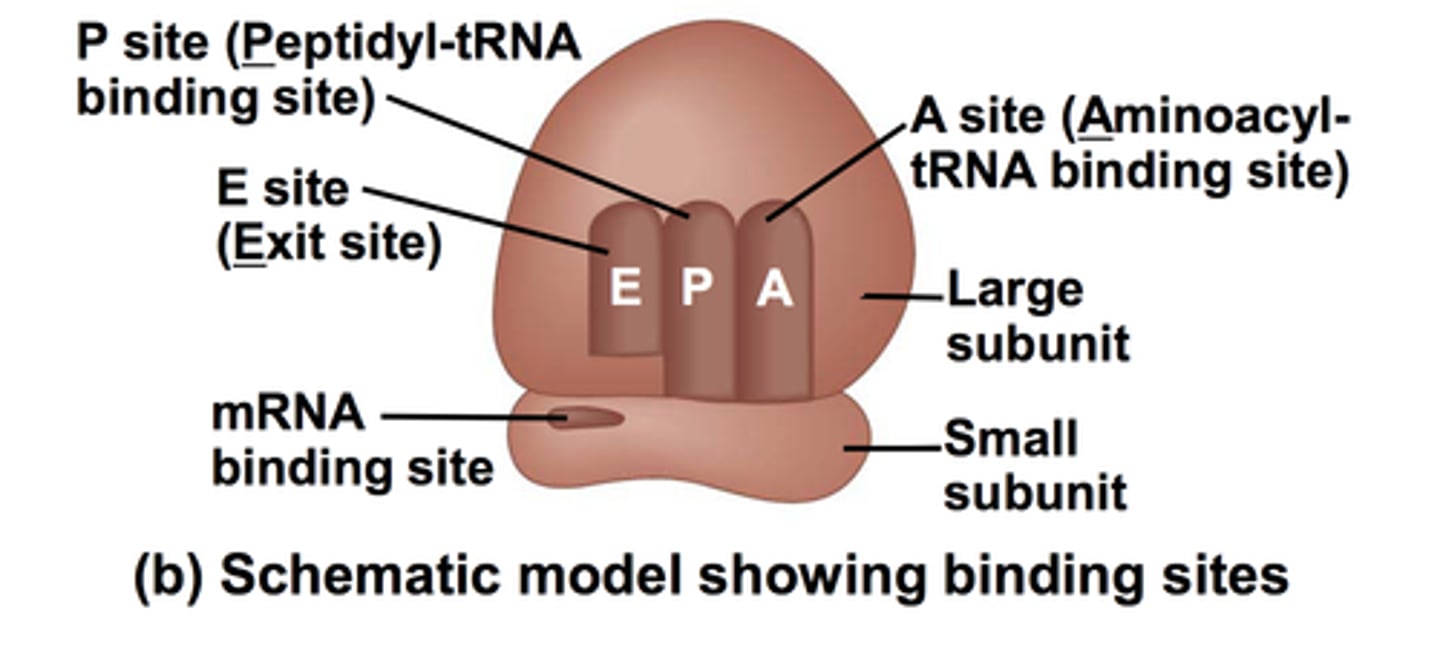
Large subunit (in ribosomes)
Has three binding sites for tRNAs: contains active site for peptide bond formation
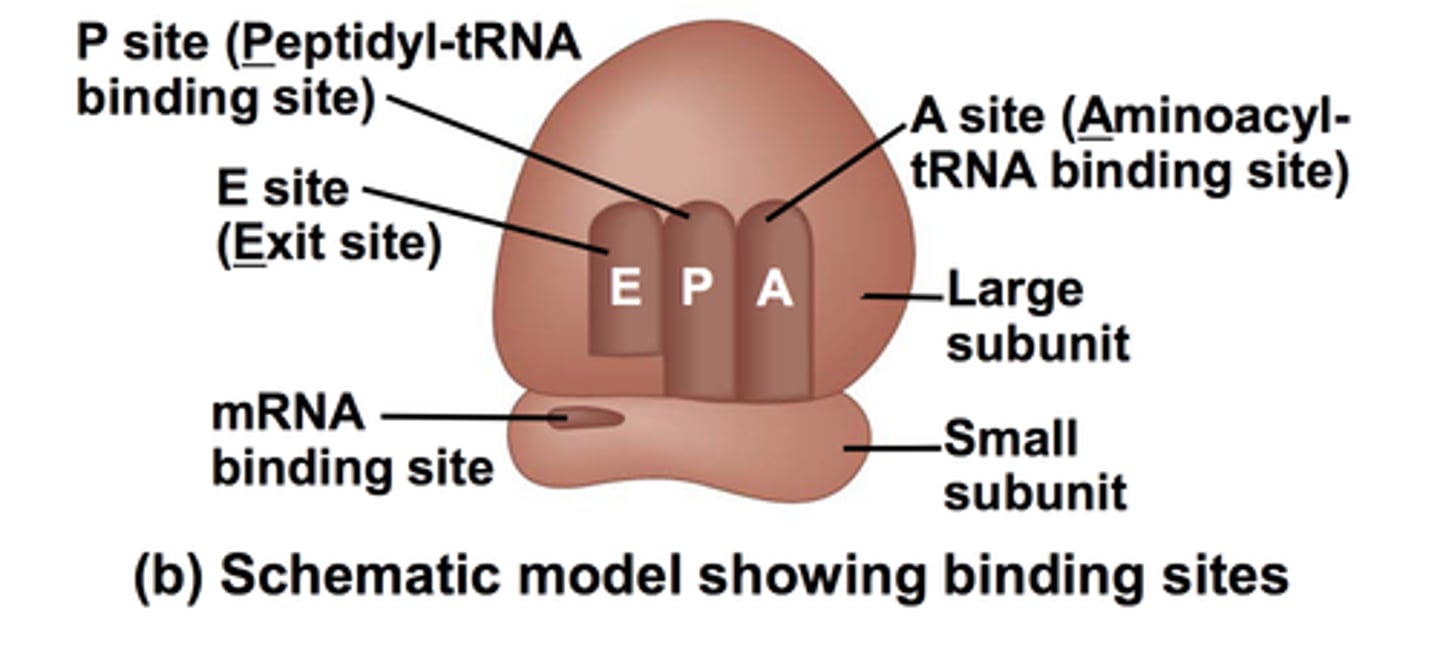
E (exit) Site
Holds tRNA that will exit (amino acid no longer attached)
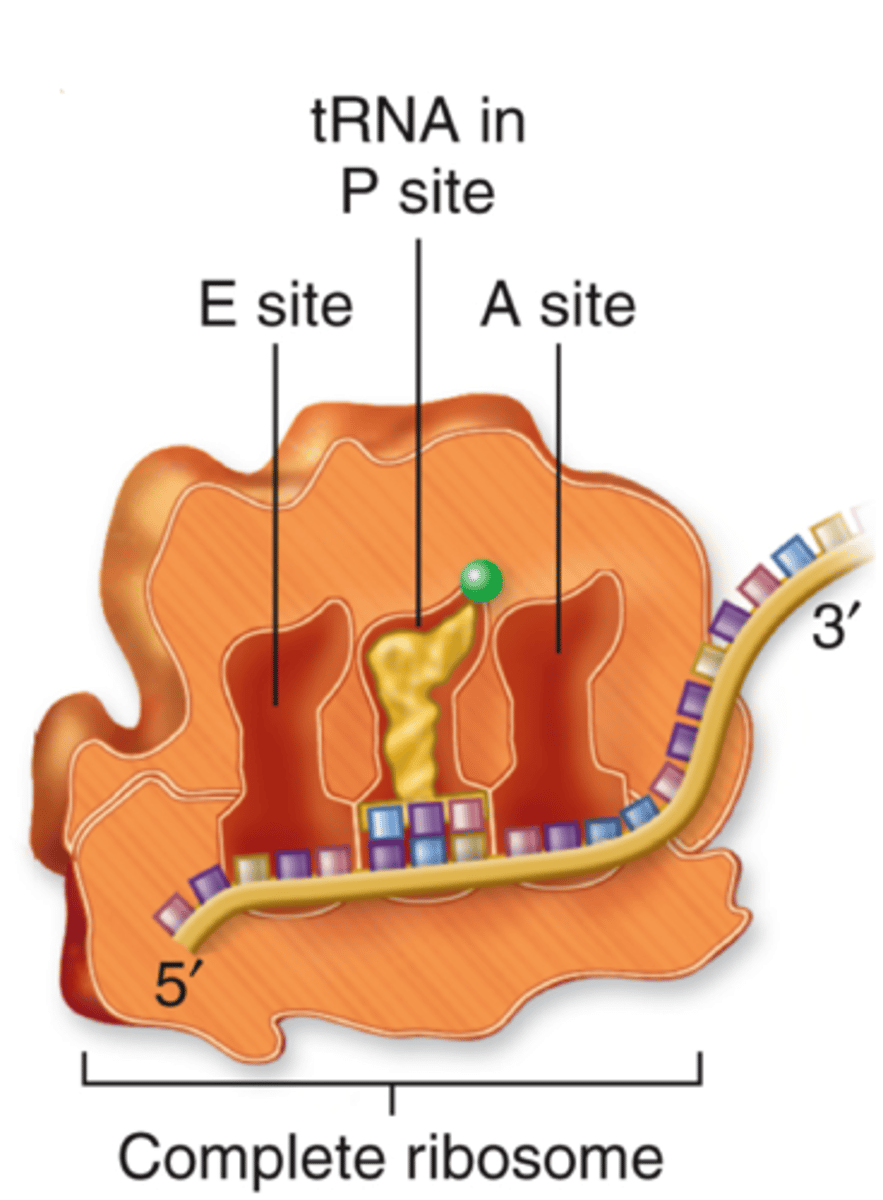
P (peptide) Site
Holds the tRNA with growing polypeptide attached
A (amino acid) Site
Holds incoming tRNA (with attached amino acid)
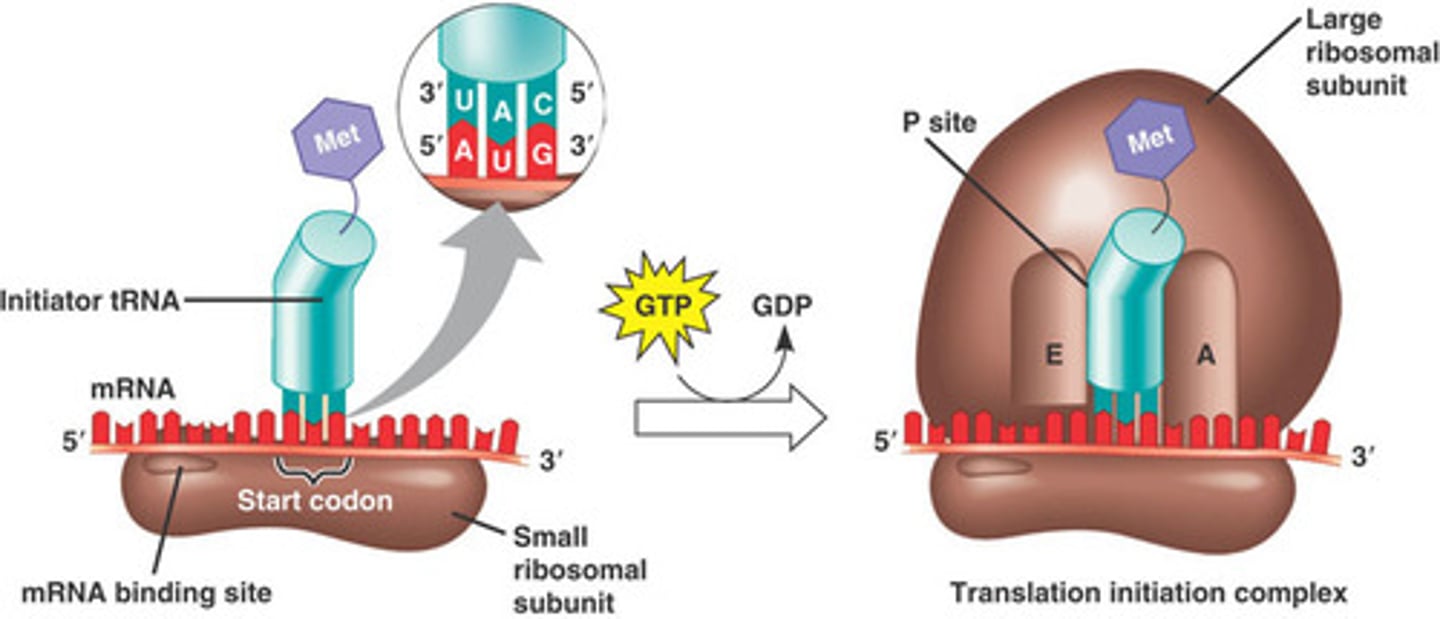
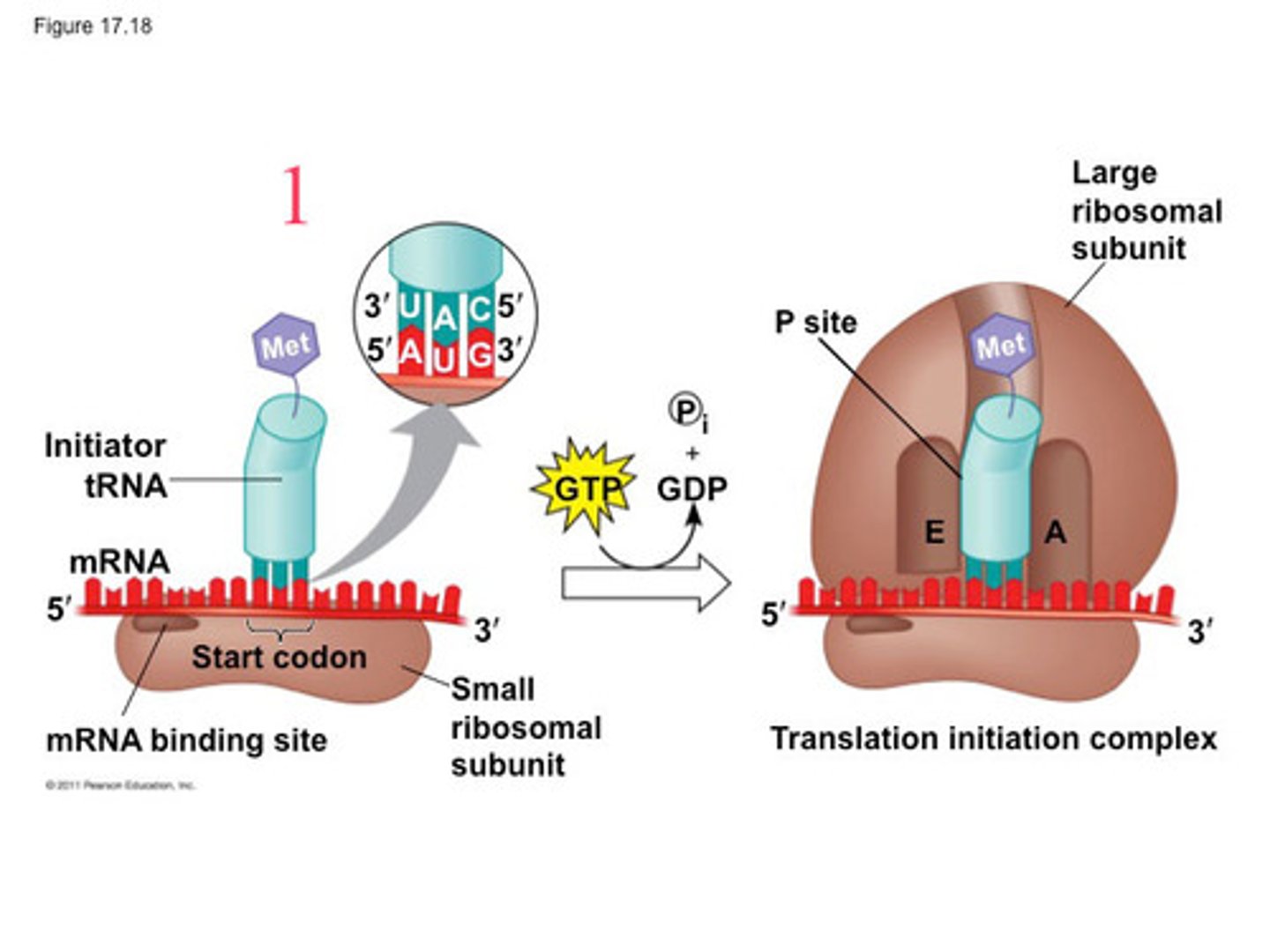
Translation initiation
1. mRNA binds to small subunit of ribosome 2. Initiator aminoacyl tRNA binds to start codon 3. Large subunit of ribosome binds, completing ribosome complex
Translation elongation
1. Arrival of tRNA/amino acid 2. Peptide-bond formation 3. Translocation
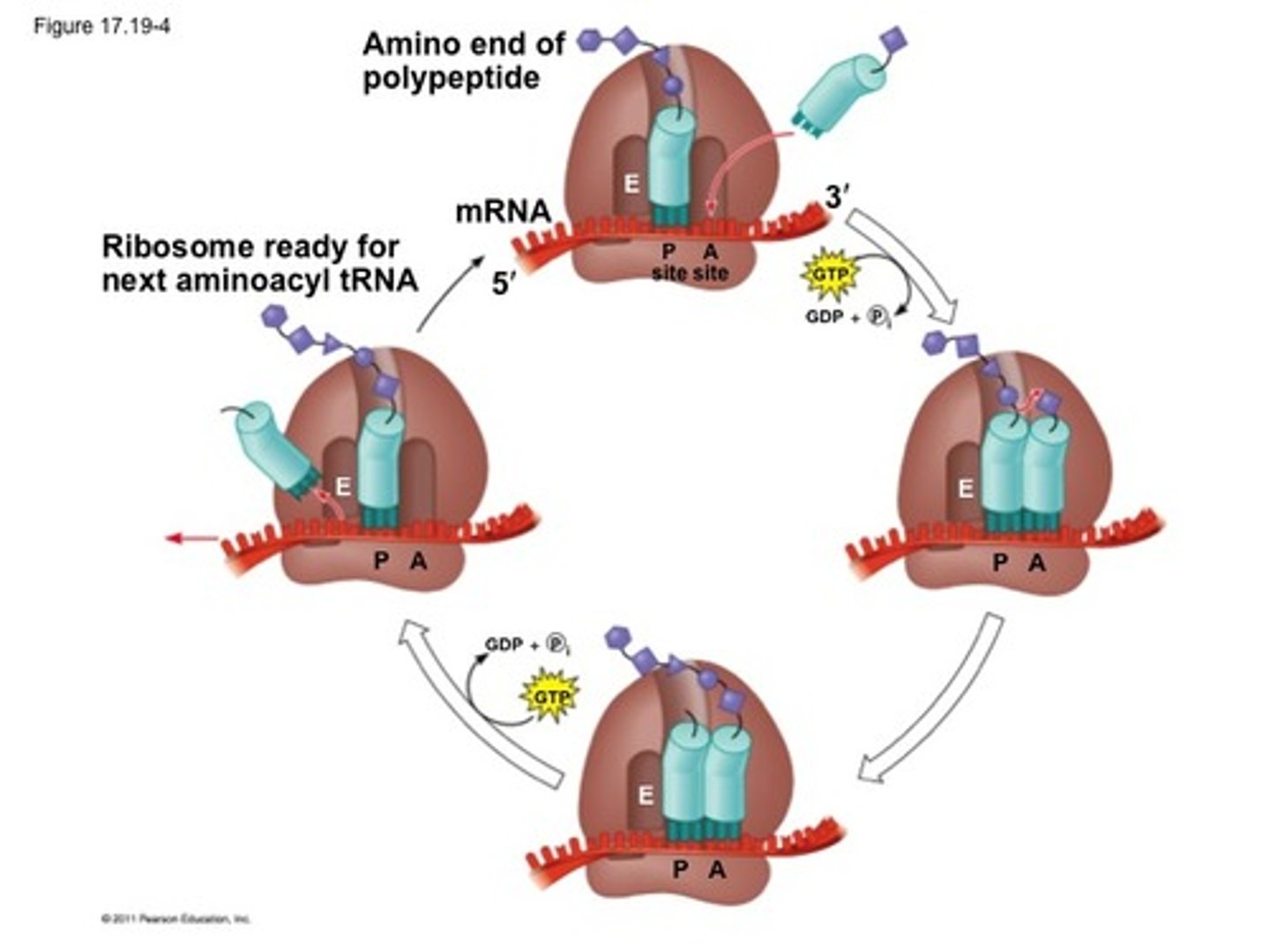
Arrival of rRNA/amino acid (translation elongation)
Appropriate tRNA (carrying an amino acid) binds to the mRNA code in the A site via complementary base pairing
Peptide bond formation (translation elongation)
Peptide chain is covalently linked to amino acid in the A site
Translocation
Ribosome moves down the mRNA (5' to 3'), moves empty tRNA into E site, moves tRNA containing polypeptide into P site, opens A site exposes new mRNA codon
What stops the translation elongation process?
When a ribosome encounters a stop codon
Translation termination
Stop codon in A site -> protein release factor enters A site, bond linking p site tRNA with polypeptide is hydrolyzed; polypeptide is released from ribosome, small and large subunit of ribosome and mRNA dissociate
In prokaryotes...
Translation begins before transcription is complete
In eukaryotes..
Transcription and translation are separated in space and time
Post-translational modification
Folding, modification (in Golgi), activation, degradation
Importance of Regulated Gene Expression
Allows cells to respond to environmental changes, improves efficiency, allows for cell differentiation
constitutively expressed genes
Genes that are needed and expressed all the time
Mechanisms of Regulation: Prokaryotes
DNA -> mRNA -> protein -> active protein
Transcriptional Control
Slow response, very efficient
Translational Control
Fast response, less efficient
Post-Translational Control
Fastest response, least efficient