27. Pathogenic and environmental fungi
1/25
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
26 Terms
S. cerevisiae mating pheromones
a-factor
α-factor
S. cerevisiae: a cells
respond to α-factor by growing a projection towards
the source of α-factor
Gene organization in the MAT, HML, and HMR loci on S. cerevisiae
chromosome III
MAT locus contains expressed mating allele, but HML and HMR contain silenced copies of a and α so that mating type switching can occur
MAT locus
region on chromosome III responsible for the expressed mating allele in S. cerevisiae.
HML locus
region on chromosome III containing silenced copies of silenced copy of MATα in S. cerevisiae.
HMR locus
region on chromosome III containing silenced copy of MATa in S. cerevisiae.
Which of the S. cerevisiae strain pairs listed below will result
in shmoo formation toward each other?
(1) two diploid cells (one which is a/α, and the other which is α/a)
(2) a haploid “a” cell that produces a-factor and expresses a
receptor that recognizes a-factor, and a haploid “α” cell that
produces α-factor and expresses a receptor that recognizes α-
factor
(3) a haploid “a” cell that produces a-factor and expresses a
receptor that recognizes α-factor, and a a/α diploid cell
(4) a haploid “a” cell that produces a-factor and expresses a
receptor that recognizes α-factor, and a haploid “α” cell that
produces α-factor and expresses a receptor that recognizes a-
factor
(5) a haploid “a” cell that produces α-factor and expresses a
receptor that recognizes α-factor, and a haploid “α” cell that
produces a-factor and expresses a receptor that recognizes a-
factor
haploid “a” cell that produces a-factor and expresses a
receptor that recognizes α-factor, and a haploid “α” cell that
produces α-factor and expresses a receptor that recognizes a-
factor
Gal4 bait and prey technique is used to determine
If a protein interacts with 2 test proteins
Gal4 bait and prey overview
create a bait construct with the Gal4 binding domain fused to your protein and two prey constructs with the Gal4 activation domain fused to each test protein; co-transform these into S. cerevisiae with a reporter gene, and if the bait and prey proteins interact, the reporter gene is transcribed
In the gal4 technique, what do you fuse the “favorite protein” to
The gal4 binding domain
In the gal4 technique, what do you fuse the non-favorite protein to
the Gal4 activation domain
In the gal4 technique, if the bait and prey interact, then what happens
the reporter gene is transcribed.
In the gal4 technique, if the bait and prey don’t interact, then what happens
the reporter gene is not transcribed.
How come most organisms have only 45unique tRNAs
the 3rd base pairs by wobble rules so that fewer codons are needed
Wobble rules
Inosine in the anticodon can pair with uracil (U), cytosine (C), or adenine (A) in the codon, allowing for flexible base pairing
G-U
Decoding CUN in S. cerevisiae
only uses two tRNAs to translate the four CUN codons
(each of which translates two codons)
Candida CUN decoding overall
Candida species translate CUG codons as serine instead of leucine
Candida CUN decoding mechanism
dedicated tRNACAGSer for CUG codons and a single tRNA IAGLeu for CUA, CUC, and CUU codon
Mycorrhiza
A symbiotic association between fungi and plant roots that enhances nutrient exchange.
endomycorrhizas
occur in ~80% of all plants. The fungal hyphae penetrate the cell walls of plant root cells and invaginate the cell membrane to create a greater contact surface area, promoting nutrient transfer
ectomycorrhizas
occur in ~10% of all plants, particularly “woody” plants (e.g., trees). The hyphae of these fungi do not penetrate individual cells within the plant root, rather they make a sheath around roots
Candida species use only two different tRNAs to
decode all four CUN codons (where N is any
nucleotide). How is this possible?
(1) the third base of an anticodon can pair by the
wobble rules (i.e., G-U, I-U, I-A, I-C)
(2) the third base of a codon shows no specificity of
binding
(3) the second base of an anticodon can pair
simultaneously with the second and third bases of a
codon
(4) the third base of an anticodon can pair by the
womble rules (i.e., G-A, I-U, I-A, I-G)
(5) the third base of an anticodon can pair by the
wobble rules (i.e., G-C, I-U, I-A, I-C)
the third base of an anticodon can pair by the
wobble rules (i.e., G-U, I-U, I-A, I-C)
Do all beer yeast make the same metabolites
NO, different strains of beer yeast produce varying metabolites, affecting flavor, aroma, and alcohol content.
Why do yeast strains differ in their alcohol tolerance
allows yeast to survive and continue fermenting in environments with high alcohol concentrations, which is crucial for producing alcoholic beverages and biofuels
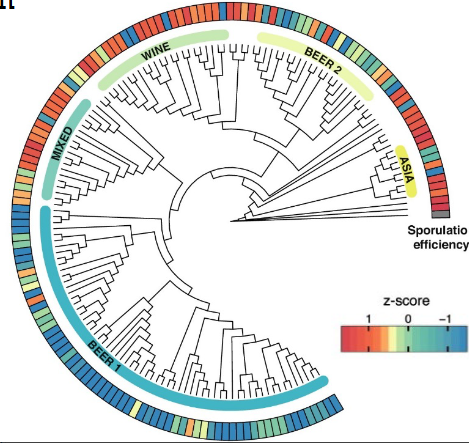
Diversity of sporulation efficiency in
industrial S. cerevisiae strains
Beer yeasts have lost their ability to effectively sporulate because they are only exposed to nutrient rich environments
Candida albicans and close relatives harbor a unique barrier to horizontal gene transfer that is not seen in other fungal species. What is this barrier?
(A) they have no cell surface glycoproteins and hence lack receptors required for viral infection
(B) they only grow as spores and hence, due to the thick spore coat, are protected against DNA entry
(C) they harbor an unusually large number of R-M complexes within the periplasm
(D) they harbor uracil, not thymine, in their DNA
(E) they use a non-standard genetic code
E. They translate CUG codons as serine (while most other organisms on the planet translate as leucine). Thus, if genes are introduced into C. albicans (let’s say from S. cerevisiae), and these genes have CUG codons in them, these will be translated as serine which may lead to unfunctional proteins