Cell bio exam 1
1/189
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
190 Terms
Spontaneous Generation
non-living objects can give rise to living organisms
Francisco Redi
Disproved spontaneous generation by showing that maggots on decaying meat came from fly eggs, not from the meat itself. Maggots on meat came from fly eggs (open container), while maggots did not grow into flies in the sealed container.
Jansen
1595, created the first compound microscope
Robert Hooke
first described cells
Antony van Leeuwenhoek
first to see prokaryotic bacteria and living protozoa/sperm/bacteria using the single lens microscope
Cell theory
cells are fundamental units of life
all living things are made of cells
new cells only arise from pre existing cells
Cells are…
small (0.1um-1mm)
membrane bound
packed with chemicals and macromolecules (gel-like)
can make copies of themselves
DYNAMIC
Molecular commonalities
DNA is genetic material
same sugars and amino acids
ATP for energy
have membrane proteins
phospholipid membranes
Central dogma
DNA → mRNA → protein
Prokaryotes
Bacteria and archaea with no nucleus or organelles, unicellular, small (0.1-10um), transcription and translation not physically separate
Eukaryotes
Animals/plants/fungi/protists that have nuclei and organelles, multicellular, large (5um-1mm), transcription and translation are physically separated
Where does eukaryotic transcription occur?
nucleus
Where does eukaryotic translation occur?
cytoplasm
Where do transcription and translation occur in prokaryotes?
cytosol
Nucleoid
where prokaryotic DNA is supercoiled, usually in a singular, circular chromosome
lysozyme
cleaves peptidoglycan in bacterial cell walls (cleaves the beta 1,4 glycosidic bond between N-acetyl muramic acid NAM and N-acetylglucosamine NAG sugars)
Organelles function in eukaryotic cells
compartmentalization of materials and reactions → regulation and efficiency (make or break things) and storage
cell/tissue differentiation: different cells have different needs/functions
SA:V
high SA:V will have…. rate of exchange of molecules with the environment relative to the internal volume than a cell with a …. SA:V
higher, low
Problems with low SA:V
slow exchange of materials with environment relative to volume (solution: endocytosis and exocytosis)
many enzymatic reactions occur on/at/in membranes
origin of eukaryotic cells
anaerobic archaea uptook aerobic proteobacterium = symbiosis of aerobic respiration
archaea are more closely related to eukarya than bacteria
true
mitochondrial endosymbiosis
all extant eukaryotes have mitochondria
plastid endosymbiosis
chloroplast is remnant cyanobacterium that allows plants to do photosynthesis
archael links
TACK archaea have proteins in common with eukaryotes, suggesting the first eukaryotic cells derived from TACK archaea
Some traits are common to all cells
ribosomes, phospholipids, nucleic acids, core metabolism
Some traits were acquired during cell evolution of specific brances and are common only to particular branches
TACK archaea: proteosome, core histones, N-linked glycans
Eukarya: genome expansion, gene duplication, multicellularity
Transition analyses
comparing metabolic pathways, cellular structures, organelles
Hypothetical model for eukaryotic cell origin
TACK archaea lose cell wall
actin cytoskeleton is altered for phagocytosis
phagocytosis of numerous bacteria by archaea with transfer of some DNA to nucleoid (horizontal gene transfer)
development of membrane around nucleoid toform nucleus and uptake of bacterial endosymbiont that maintains independence as mitochondrion
mitochondria multiply in primitive eukaryotic cells
Gene families
Homologs
genes that are related by descent, with similar sequences and functions
ex. orthologs (different species, same function) AND paralogs (same species, same function)
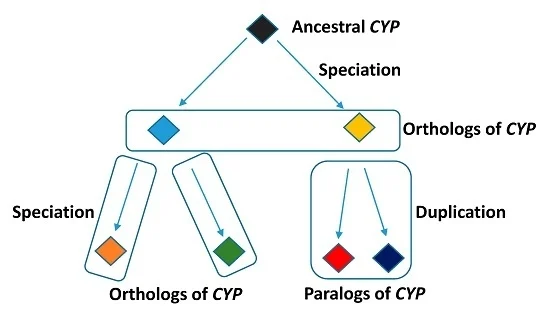
Paralogs
arise by a gene duplication event within a single genome, go on to have many individual organisms with multiple copies of genes with similar sequences/functions
same species, same function
Orthologs
different species, same function
Evolution of genes and genomes
intragenic mutation
gene duplication (meiosis) → functional divergence (one can develop mutations)
DNA segment shuffling
horizontal DNA transfer (antibiotic resistant genes or endosymbiosis)
Cell lysis
put tissue in buffer at specific pH
disrupt tissue w/mortar and pestle or blender or homogenizer
French press
put cells under high pressure → rapid release of pressure
Sonification
probe vibrates very rapidly
cavitation
vapor filled cavities form and collapse to lyse cells
Detergent lysis
dissolves cell membrane (disadvantage: can denature protein, doesn’t work on cells with cell wall)
Osmotic shock
put cells in hypotonic solution (water flows in and cell lyses)
Cell fractionation (centrifugation)
differential, density gradient
differential centrifugation
low spins, separation based on size/density of subcellular structures (pellet and supernatent); separate cytosol from rest of cell
density gradient ultracentrifugation
high speed
velocity sedimentation
sucrose gradient and lysate on top → centrifuge → small less dense things on top, larger denser things near bottom
equilibrium sedimentation
lysate is mixed into gradient → centrifuge → cell components migrate according to density until neutral density is reacted
Protein purification
chromatography
Gel filtration (size exclusion)
separation by SIZE
solvent flows through porous beads and small proteins get stuck in beads (big proteins elute first)
Ion exchange
separation based on CHARGE
any protein can be positively or negatively charged, depends on buffer pH and isoelectric point pI
isoelectric point pI
pH where protein is neutral (no net charge)
pH > pI
protein is negatively charged (fewer protons)
pH < pI
protein is positively charged
Anion exchange
positively charged beads → negatively charge molecules interact while positively charged molecules flow through (positively charged proteins elute first)
increase salt concentration
disrupt proteins that interact with positive matrix
How does phosphorylation affect charge?
adds negative charge to proteins
Cation exchange
uses negatively charged matrix so positively charged molecules interact
Affinity chromatography
stationary phase has something covalently attached; protein sticks to something specifically while other proteins flow through (low affinity); eluting solution contains something that compete for binding
SDS page
determine purity
vertical polyacrylamide gel electrophoresis (negative cathode → positive anode) → negatively charged proteins migrate towards positive pole (bottom) → small things move quickly, slow large things move slowly
SDS
sodium dodecyl sulfate; strong anionic detergent; denatures proteins AND gives them net negative charge; all proteins get same mass/charge ratio, separation on SIZE only
What does SDS PAGE tell us?
size, purity, how much (darker band = more)
beta mercaptoethanol
breaks disulfide bonds
2D SDS PAGE
first dimension is isoelectric focusing (proteins separated by electrophoresis in gel w/pH gradient, proteins migrate until they reach a place w/no net charge pI)
second dimension is SDS PAGE
How does phosphorylation affect isoelectric point (pI)?
pI lowers because phosphorylation adds more negative charge therefore more acidic conditions (more H+ ions) are needed to reach pI
Antibodies
produced in B cells in response to antigens
polyclonal antibodies
recognized different epitopes of the same antigen
monoclonal antibodies
normal, somatic cells can be fused with tumor cells to form a heterokaryon; recognize a single epitope
Western Blotting
detection of a protein on a membrane after SDS PAGE
primary antibodies
recognize protein of interest (produced in one species)
secondary antibodies
recognize first antibody (produced in another species and recognizes the Fc region of the first species’ IgG)
contains label (enzyme)
2 identical SDS PAGE gels are run
one gel stained for total protein, one gel blotted onto membrane and investigated for single protein w/an antibody
Protein interaction methods
use genetic engineering to add “epitope” tag (extra protein [fusion protein] sequence that is translated w/gene for protein)
Tags can be used for…
affinity purification and ‘pull down’ assays
Immunoprecipitation assays
detection (Western blot, microscopy)
Pull down assays
depends on affinity chromatography → protein binds to column via glutathione S transferase as “bait” ; proteins from cell lysate are added “prey” ; bait and prey complexes eluted from column together
Co-immunoprecipitation
harvest and lyse cells/tissue
inert bead w/specific antibody will bind to protein (+ other interacting proteins)
elute protein complex from beads
Chromatin immunoprecipitation (ChIP)
protein DNA interactions
Cross link transcription factors of proteins (w/formaldehyde)
lyse cells
break DNA into small fragments
use antibodies against protein
protein is purified along w/the bound DNA
amplify the purified DNA (DNA corresponds to transcription regulators)
Resolution
smallest distance b/w two points in an object that is still perceived as separate in the image
highest resolution of human eye
0.2mm (200 um)
highest resolution of light microscope
200 nm (0.0002 mm)
highest resolution of electron microscope
0.2 nm
standard compound light microscopes
upright
inverted: cells in petri dishes → bottom of dish is ‘cover slip’, image from bottom
effective magnification
eyepiece (10x) x objective lens (up to 100x)
Cells in tissues
fix: heat w/chemicals to maintain morphology
section: cut into thin slices w/microtome (embed in wax or plastic) → slice think sections
place section on slide (maybe stain)
image
Problem of light microscopes
light microscope have max. resolution of 0.2 um (200nm) and many cellular structures are smaller than this
Solution
use electrons (very short wavelengths), refracted by smaller structures
Transmission electron microscopy (TEM)
very high resolution, 2nm
can only image fixed and sectioned samples
similar to compound light
electrons pass through very thin slice of specimen
Scanning electron microscopy (SEM)
very high resolution, 3-20nm
image intact tissues/cells but can’t see inside (unless broken open)
similar to dissecting microscope; electrons bounce off specimen
look at cell surface (vs. slice)
Eukaryotic chromosome structure
multiple linear chromosomes per nucleus, each chromosome is a single DNA molecule that maintains genetic info and replicates genetic info
Prokaryotic chromosome structure
one circular chromosome (nucleoid) and plasmids (extra chromosomal DNA)
telomeres
protective caps on chromosome, get shorter every time cell divides
centromere
mostly heterochromatin, required for segregation by kinetochores in M-phase
Nuclear envelope
boundary of nucleus; 2 lipid bilayers (inner and outer, contiguous with ER)
nuclear pore
gated, control entry and exit of macromolecules (RNA and proteins)
nucleolus
where ribosomes are made; transcription of rRNAs by RNA polymerase I and ribosome assembly
nuclear lamina
meshwork of supportive proteins
Chromosomes are made from chromatin
DNA and associated proteins
nucleosome
core unit, DNA wraps around histone heterooctamer core (~1.6x)
linker DNA
variable length, depends on chromatin state
nuclease-protection assays
isolate nuclei with DNase I (naked DNA is cut)
treat with high conc. of salt to separate histone octamer and DNA
DNA gel electrophoresis - big DNA is more negatively charged and stays near the top, while positively charged small DNA moves to the bottom
Histones
heterooctamer- 2 of each protein subunit (H2A, H2B, H3, H4)
subunits are small, basic (lysine and arginine) with a positive charge, and have N-terminal tails that can be modified
Histone H1
not part of core, helps compact DNA in heterochromatin (mitotic chromosomes)
DNA folding
naked DNA —> nucleosome beads on a string —> 30nm fiber of chromatin —> loops —> mitotic chromosomes
Euchromatin
loose, long linker DNA between nucleosomes = express genes