Biology Test 3
0.0(0)
0.0(0)
Card Sorting
1/115
There's no tags or description
Looks like no tags are added yet.
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
116 Terms
1
New cards
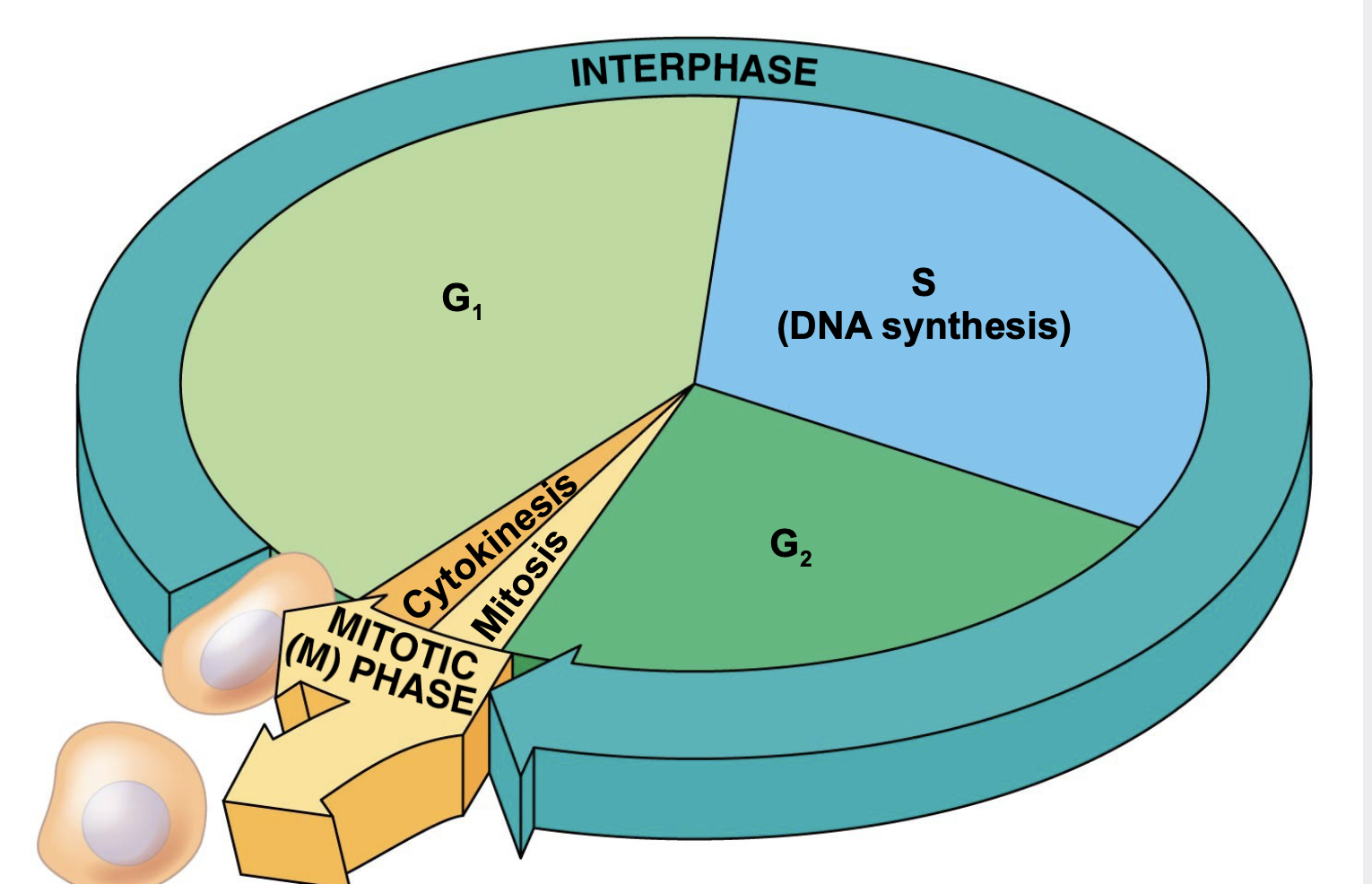
3 stages of cell cycle and description
1. interphase- periods (phases) of growth and DNA replication:
G1 phase - primary growth phase
S phase - DNA replication phase \n G2 phase - second growth phase
2. nuclear division (M phase)- separation of the replicated DNA into daughter nuclei. There are two forms of nuclear division: mitosis and meiosis
3. cytokinesis- division of the cytoplasm and other parts (organelles) of the cell
2
New cards
Most of a cell’s lifetime is spent in
Interphase
3
New cards
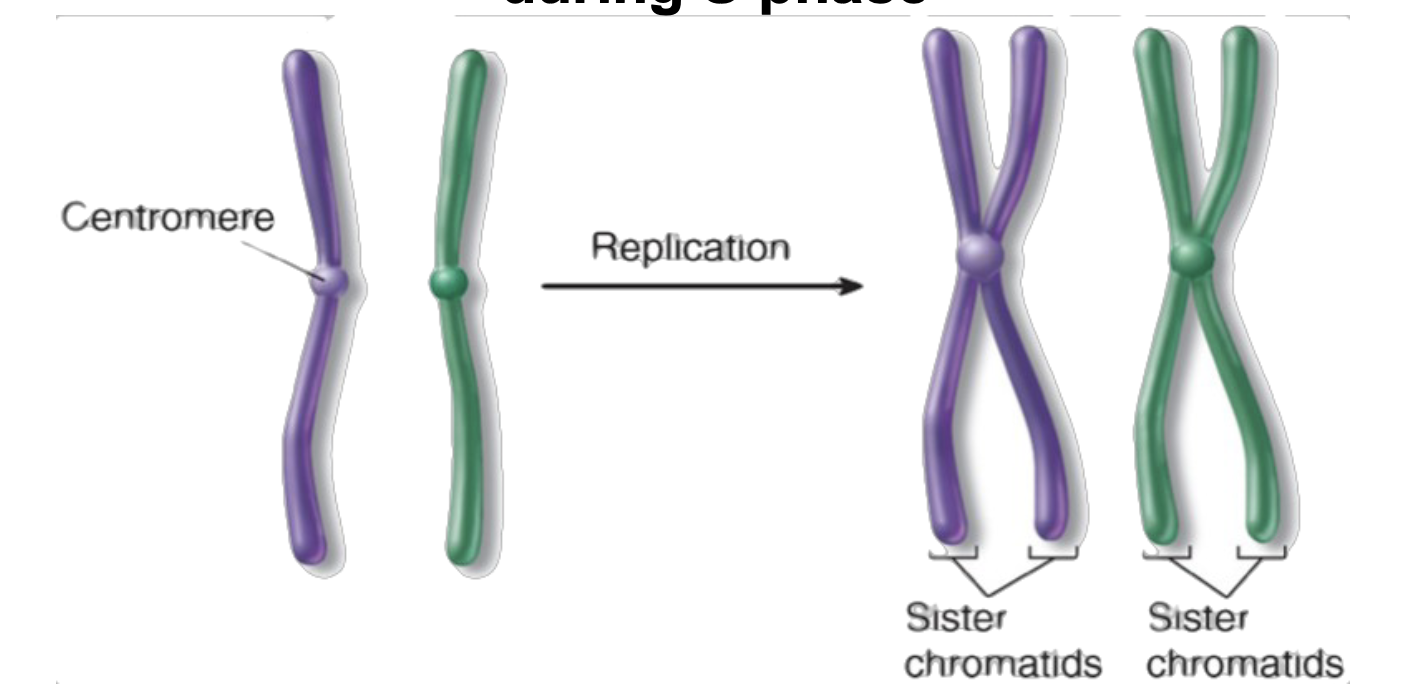
sister chromatids
Duplicated chromosomes during S phase that are held together by a protein complex at a spot in the center of the chromosome called the centromere
4
New cards
Mitosis
produces two daughter cells that are genetically identical to the parent cell (and each other). Divided into prophase, metaphase, anaphase, and telophase
5
New cards
Meiosis
produces sex cells (sperm/egg) for sexual reproduction that have only half the chromosomes as the parent cell
6
New cards
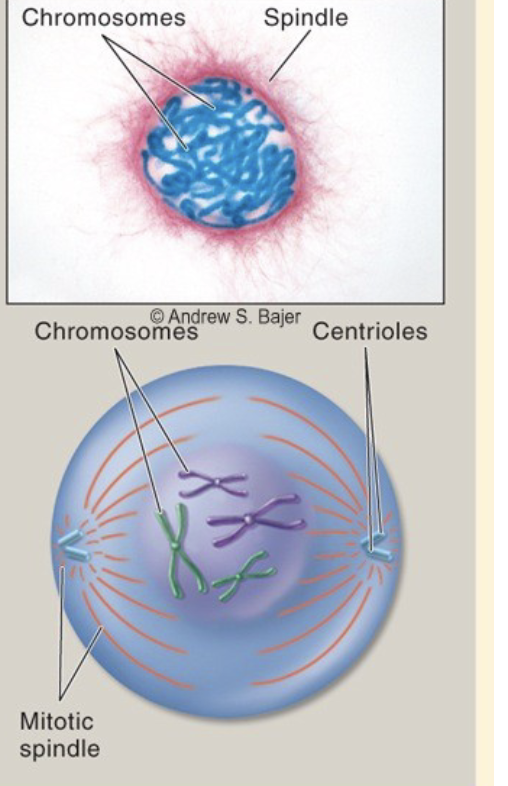
Prophase
.chromosomes condense (and become visible with a light microscope) \n .the nuclear envelope breaks down \n .the cytoskeleton begins to assemble a network of protein cables called the spindle \n -two spindles form such that they are positioned on opposite ends of the cell (poles) \n -for each set of sister chromatids, one sister chromatid attaches to one spindle, and the other sister chromatid attaches to the opposite spindle
7
New cards
histone and chromatin
Inside the nucleus, the chromosomal DNA is wound around histone proteins.
DNA bound by proteins is called chromatin.
DNA bound by proteins is called chromatin.
8
New cards
Prophase and Prometaphase
prophase includes:
. spindle formation and \n . the beginning of chromosome condensation,
prometaphase includes: \n • completion of chromosome condensation \n • attachment of chromosomes to spindle \n • breakdown of nuclear envelope
\
. spindle formation and \n . the beginning of chromosome condensation,
prometaphase includes: \n • completion of chromosome condensation \n • attachment of chromosomes to spindle \n • breakdown of nuclear envelope
\
9
New cards
Metaphase
the chromosomes are aligned in the center of the cell by the spindle. The spindle fibers are attached kinetochore on opposite sides of centromeres.
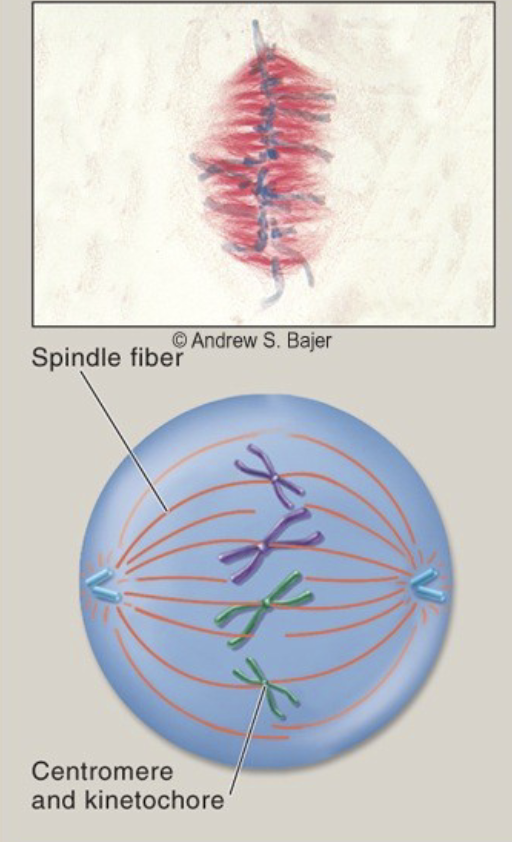
10
New cards
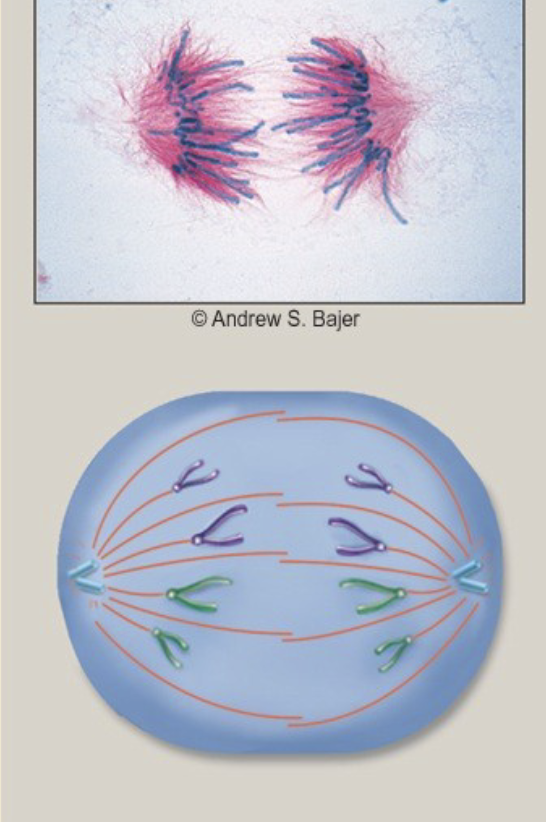
Anaphase
.sister chromatids separate from one another \n .microtubules pull the sister chromatids toward opposite poles \n -this places one copy of each chromosome at each poles
11
New cards
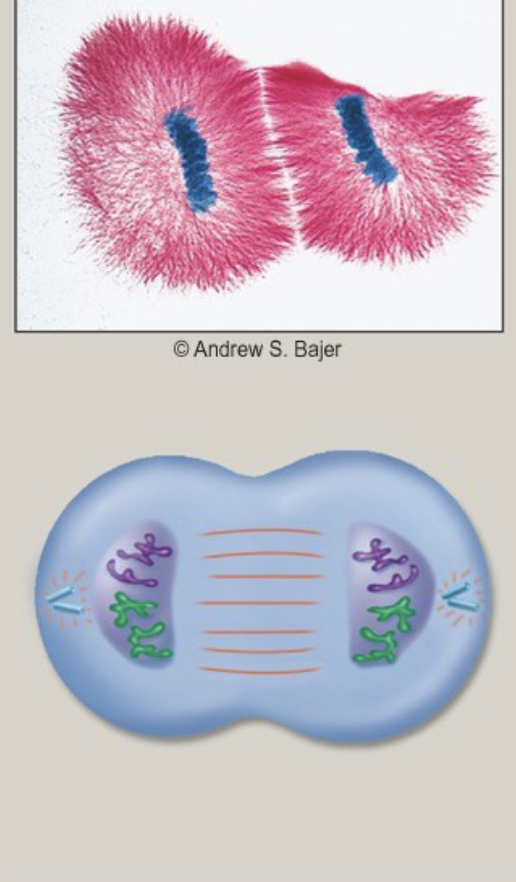
Telophase
.the spindle is dismantled
\-a nuclear envelope forms around each set of chromosomes at each pole
.the chromosomes begin to decondense
\-essentially, the cell is forming the two new nuclei
\-a nuclear envelope forms around each set of chromosomes at each pole
.the chromosomes begin to decondense
\-essentially, the cell is forming the two new nuclei
12
New cards
Cytokinesis
Cytokinesis is the division of the cytoplasm into roughly equal halves.
.In animals: occurs by protein filaments that contract and pinch the cell in two. This action is evident as a cleavage furrow that appears between the daughter cells.
.In Plants: a new plasma membrane and cell wall is laid down to divide the two daughter cells. The forming cell wall is called the cell plate
.In animals: occurs by protein filaments that contract and pinch the cell in two. This action is evident as a cleavage furrow that appears between the daughter cells.
.In Plants: a new plasma membrane and cell wall is laid down to divide the two daughter cells. The forming cell wall is called the cell plate
13
New cards
Gametes
reproductive cells (eggs and sperm). The formation of gametes must involve some mechanism to halve the number of chromosomes found in somatic cells. if not, the number of chromosomes would double with each fertilization
14
New cards
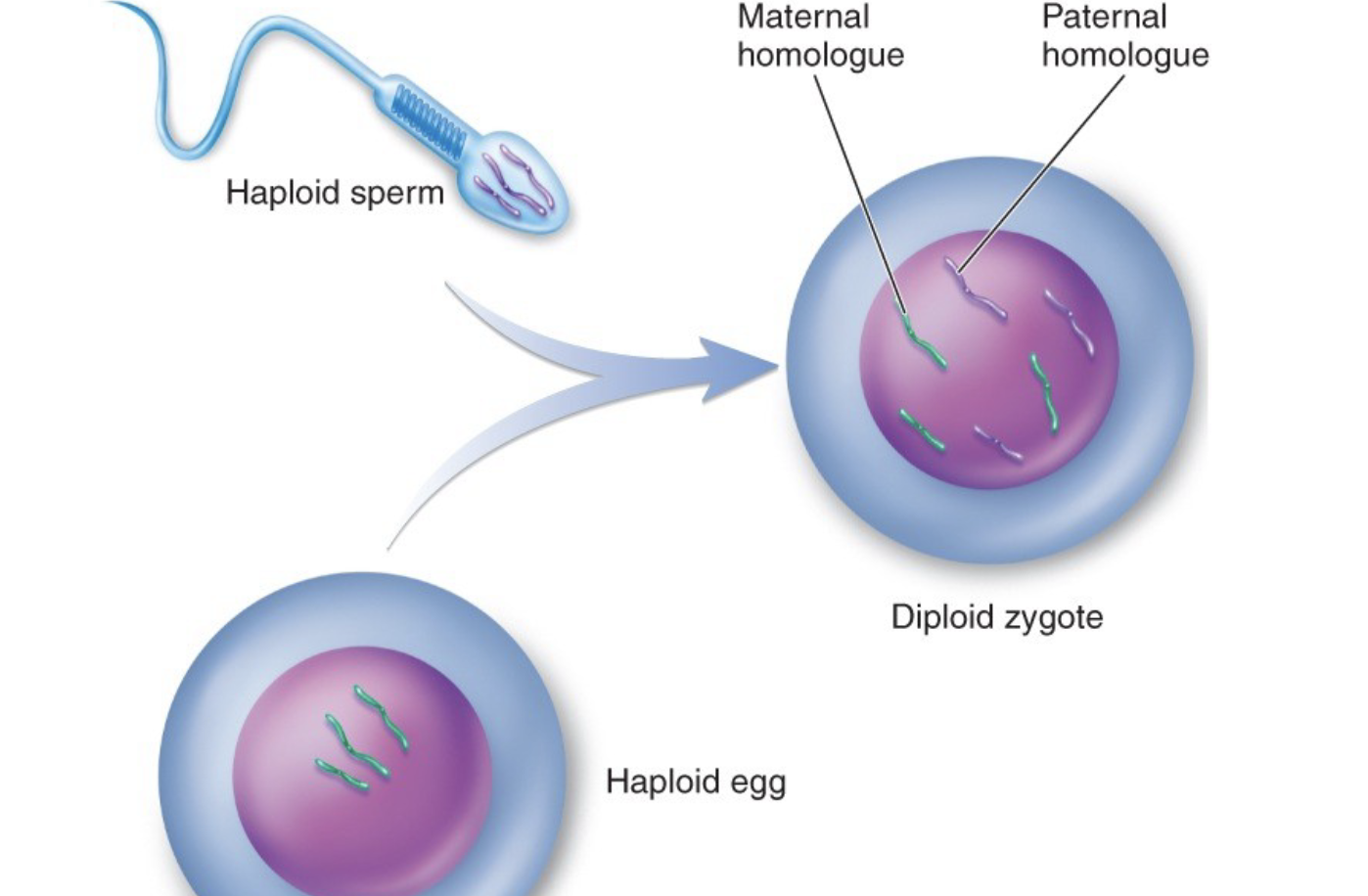
meiosis in gametes
the process of nuclear division used in forming gametes to halve the chromosome number
15
New cards
Somatic Cells and Diploids
.Somatic cells (“regular” cells of the body not involved in reproduction) have two sets of chromosomes and are called diploid.
• These two sets of chromosomes exist as homologous pairs of chromosomes.
• homologous pair = two versions of the same type of chromosome (not duplicated chromosomes).
• Homologous chromosomes contain the same kinds of genes, but different versions of those genes may be on the two different chromosomes in a homologous pair.
• These two sets of chromosomes exist as homologous pairs of chromosomes.
• homologous pair = two versions of the same type of chromosome (not duplicated chromosomes).
• Homologous chromosomes contain the same kinds of genes, but different versions of those genes may be on the two different chromosomes in a homologous pair.
16
New cards
Gametes and Haploids
Gametes have only one set of chromosomes and are said to be haploid. \n • Gametes have one chromosome from each homologous pair.
17
New cards
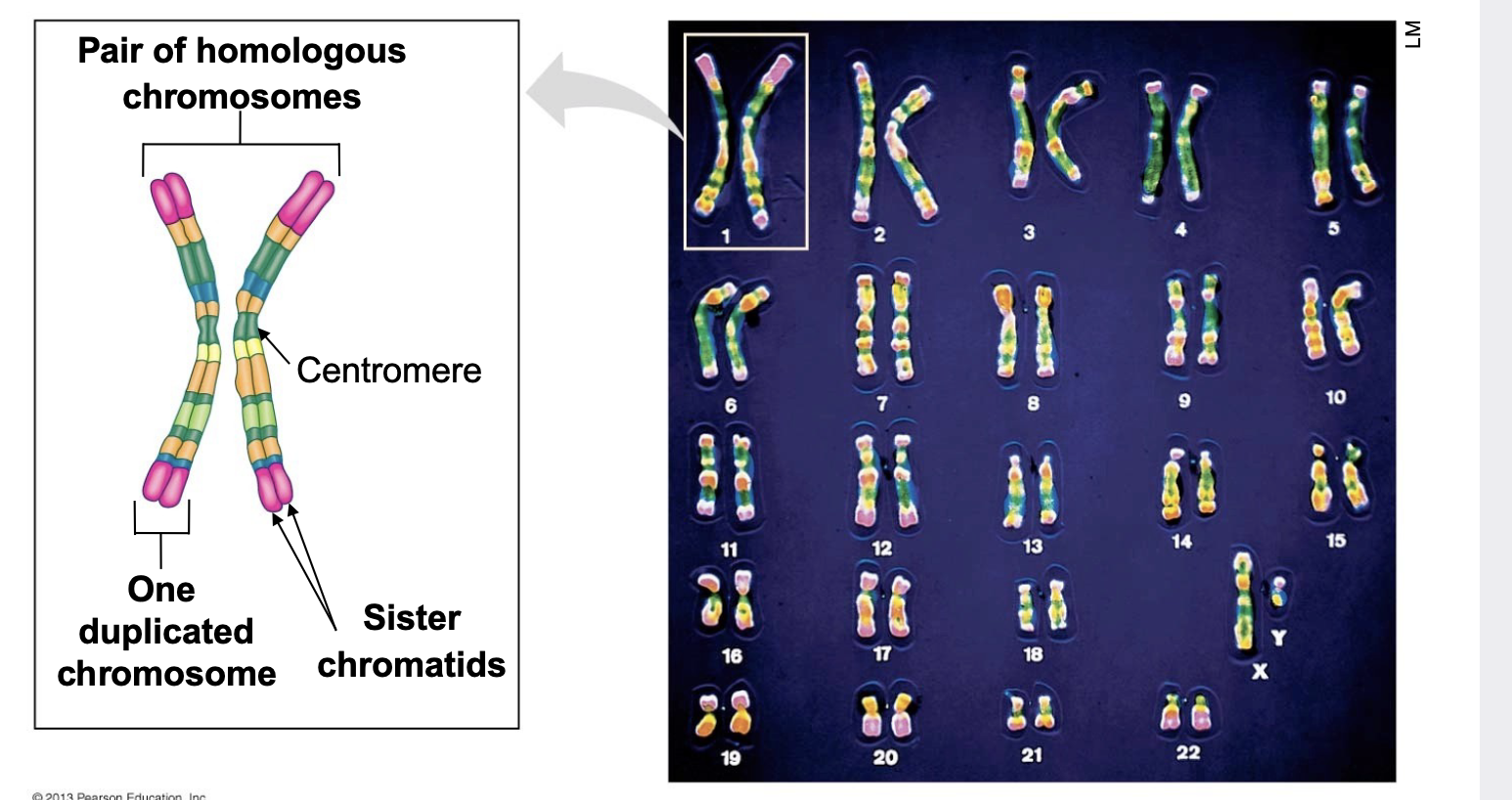
How many pairs of human chromosomes? and description
23
1. sister chromatids - exact duplicates of a chromosome that are produced only when a cell is going to divide
2. homologous chromosomes -non-identical pairs of chromosomes that contain the same kinds of genes on each. You inherit one chromosome in each pair from your mother and the other chromosome in each pair from your father when the egg and sperm fertilize.
1. sister chromatids - exact duplicates of a chromosome that are produced only when a cell is going to divide
2. homologous chromosomes -non-identical pairs of chromosomes that contain the same kinds of genes on each. You inherit one chromosome in each pair from your mother and the other chromosome in each pair from your father when the egg and sperm fertilize.
18
New cards
diploids and haploids in humans
. diploid cells contain 22 pairs of somatic chromosomes and one pair of sex chromosomes (23 pairs = 46 chromosomes total in diploid human cells)
.haploid cells contain 22 total somaitc chromosomes and one sex chromosome (no pairs; 23 chromosomes total in haploid human cells)
.haploid cells contain 22 total somaitc chromosomes and one sex chromosome (no pairs; 23 chromosomes total in haploid human cells)
19
New cards
Meiosis differs from mitosis in that
• Meiosis separates the pairs of homologous chromosomes so that one homolog goes into one daughter cell and the other homolog goes into a different daughter cell. (Mitosis does not separate homologous pairs.)
• Meiosis involves two nuclear divisions, producing four haploid cells. (Mitosis involves one division to produce two diploid cells) Meiosis is thus used to produce haploid gametes, so that fertilization restores the diploid number of chromosomes.
• Meiosis involves two nuclear divisions, producing four haploid cells. (Mitosis involves one division to produce two diploid cells) Meiosis is thus used to produce haploid gametes, so that fertilization restores the diploid number of chromosomes.
20
New cards
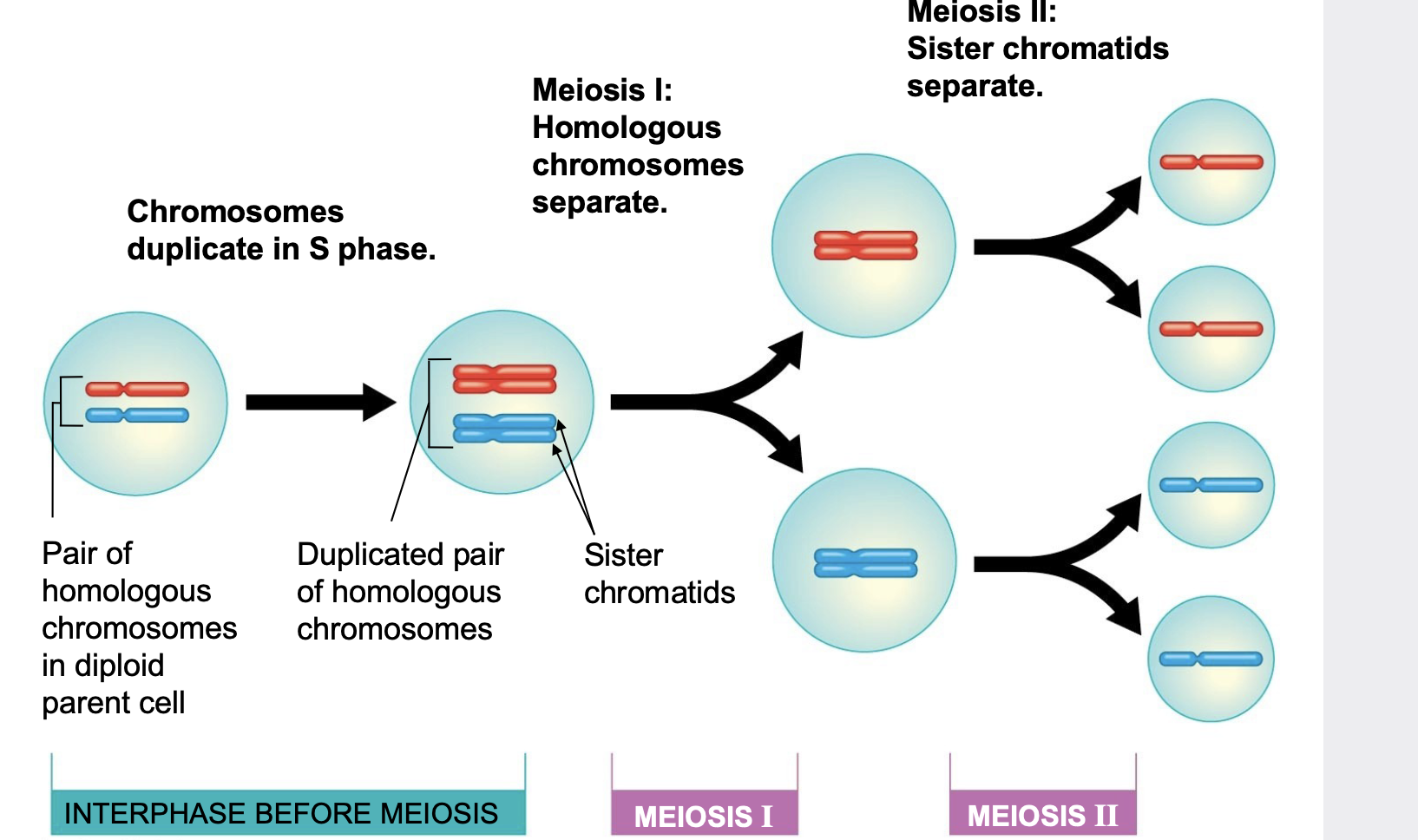
Mitosis vs Meiosis Division
.Mitosis is one division. sister chromatids are separated into diploid daughter cells.
.during meiosis I, homologous chromosomes (still in the form of sister chromatids) are separated into daughter cells. In meiosis II, sister chromatids are separated into daughter cells (meiosis II is similar to mitosis)
.during meiosis I, homologous chromosomes (still in the form of sister chromatids) are separated into daughter cells. In meiosis II, sister chromatids are separated into daughter cells (meiosis II is similar to mitosis)
21
New cards
Stages of Meiosis
Meiosis I:
1\. Prophase I \n • homologous chromosomes pair up \n 2. Metaphase I \n • the paired homologous chromosomes align on a \n central plane \n 3. Anaphase I \n • homologues separate and move to opposite poles
4\. Telophase I \n • chromosomes gather at each of the two poles
\
Meosis II:
1\. Prophase II \n • new spindle forms to attach to chromosomes \n 2. Metaphase II \n • chromosomes align along a central plane \n 3. Anaphase II \n • sister chromatids separate and are pulled to opposite \n poles \n 4. Telophase II \n • the nuclear envelope is reformed around each of the four sets of daughter chromosomes
1\. Prophase I \n • homologous chromosomes pair up \n 2. Metaphase I \n • the paired homologous chromosomes align on a \n central plane \n 3. Anaphase I \n • homologues separate and move to opposite poles
4\. Telophase I \n • chromosomes gather at each of the two poles
\
Meosis II:
1\. Prophase II \n • new spindle forms to attach to chromosomes \n 2. Metaphase II \n • chromosomes align along a central plane \n 3. Anaphase II \n • sister chromatids separate and are pulled to opposite \n poles \n 4. Telophase II \n • the nuclear envelope is reformed around each of the four sets of daughter chromosomes
22
New cards
nondisjunction
Mistakes during meiosis, the members of a chromosome pair fail to separate
– producing gametes with an incorrect number of chromosomes.
– after fertilization of such a gamete, the zygote will have the incorrect number of chromosomes.
• Nondisjunction can occur during meiosis I or II.
. Zygotes with abnormal chromosome number will usually not develop or will have abnormalities.
– producing gametes with an incorrect number of chromosomes.
– after fertilization of such a gamete, the zygote will have the incorrect number of chromosomes.
• Nondisjunction can occur during meiosis I or II.
. Zygotes with abnormal chromosome number will usually not develop or will have abnormalities.
23
New cards
Trisomy 21
example of nondisjunction where a person receives three copies of chromosome 21.
• The resulting condition is called Down syndrome.
• The resulting condition is called Down syndrome.
24
New cards
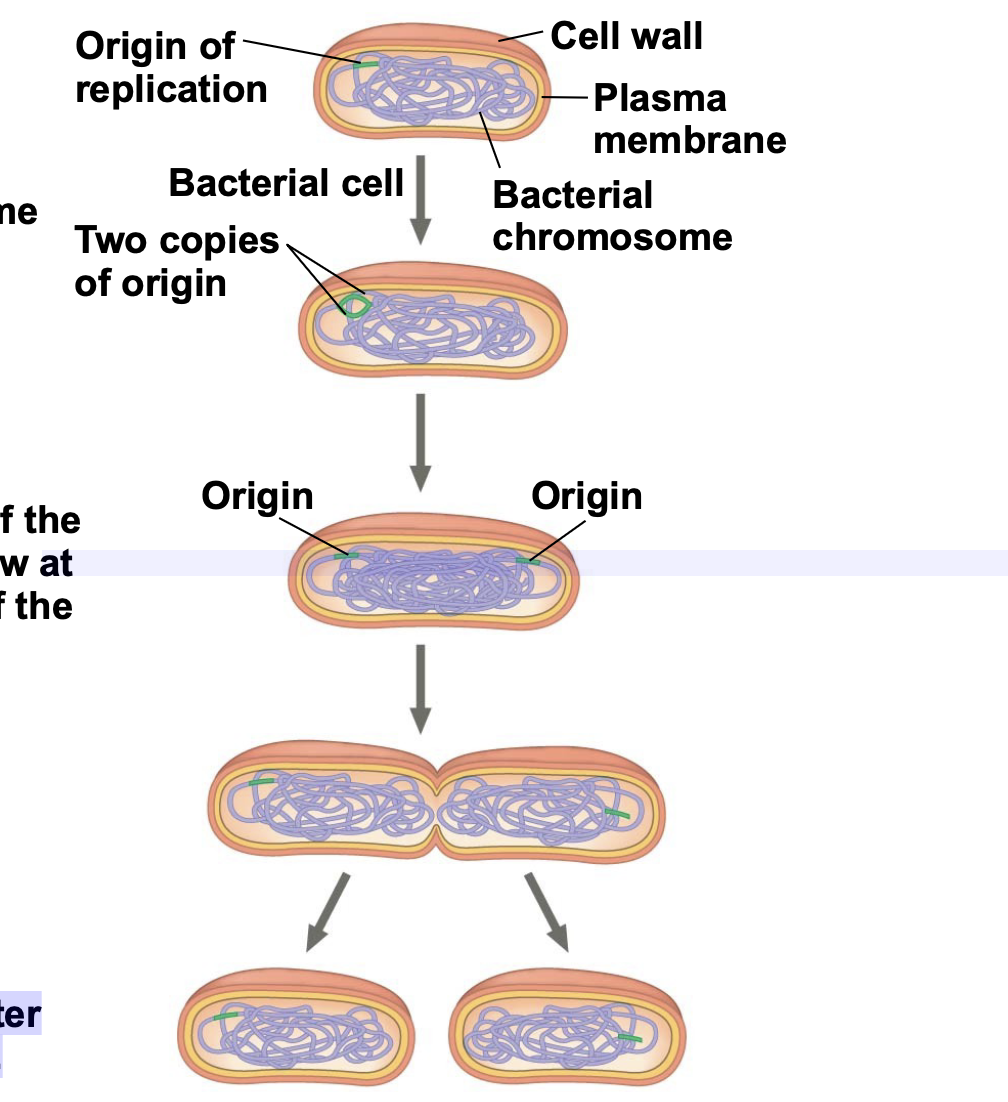
Binary Fission in Bacteria
.Prokaryotes (bacteria and archaea) do not have a cell cycle that undergoes mitosis/meiosis. They reproduce by a type of cell division called binary fission.
\-In binary fission, the chromosome replicates (beginning at the origin of replication), and the two daughter chromosomes actively move apart.
. The plasma membrane pinches inward, dividing the cell into two
\-In binary fission, the chromosome replicates (beginning at the origin of replication), and the two daughter chromosomes actively move apart.
. The plasma membrane pinches inward, dividing the cell into two
25
New cards
Binary fusion Stages
1. Chromosome replication begins.
2. One copy of the origin is now at each end of the cell.
3. Replication finishes.
4. Two daughter cells result.
26
New cards
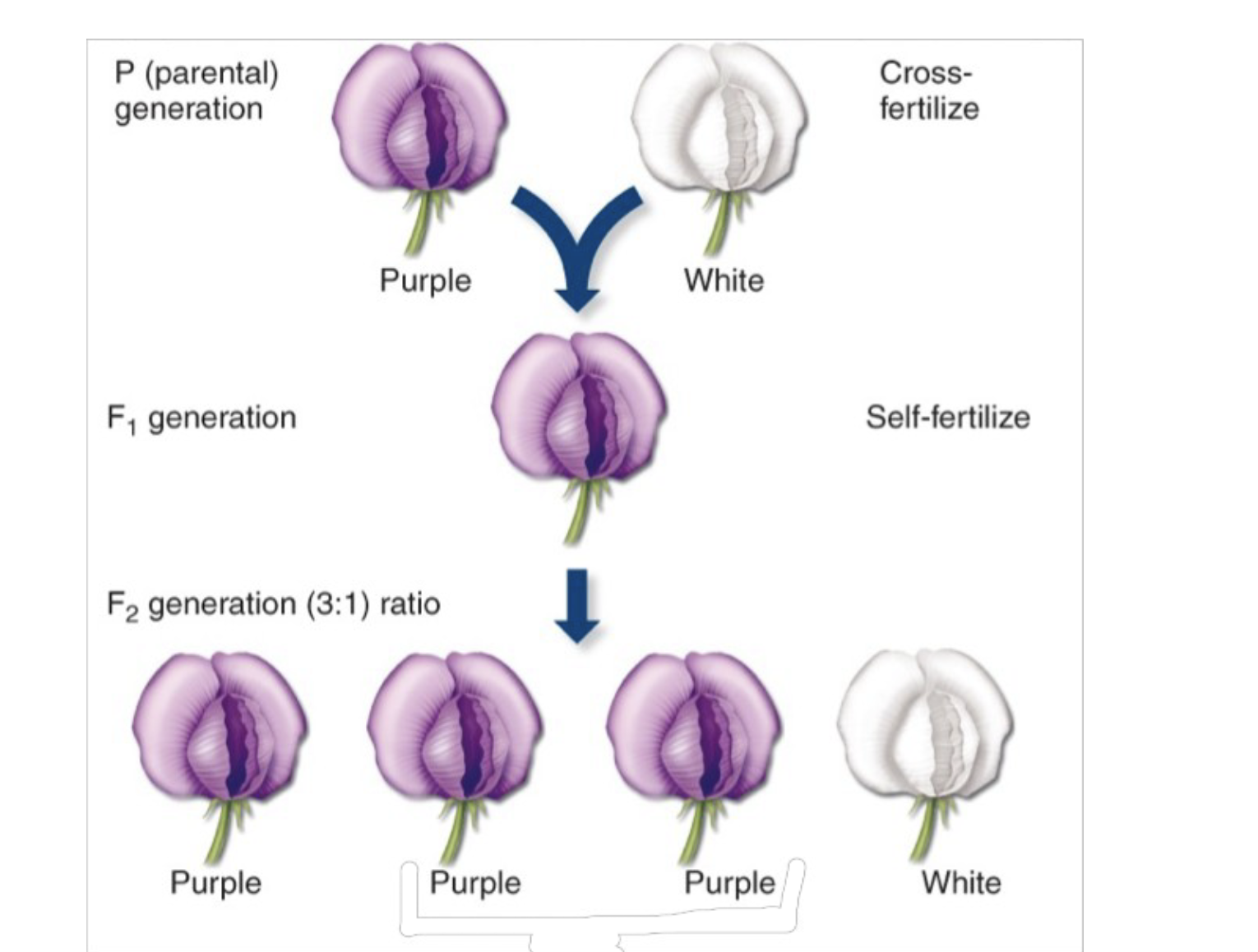
monohybrid cross
.The type of cross that led Mendel to realize traits were heritable. It is a cross between purebred parent plants that differ in only one character.
.Hybrids are the offspring of two different \n purebred varieties. \n – The parental plants are called the P generation. \n – Their hybrid offspring are called the F1 generation. \n – A cross of the F1 plants forms the F2 generation.
.Hybrids are the offspring of two different \n purebred varieties. \n – The parental plants are called the P generation. \n – Their hybrid offspring are called the F1 generation. \n – A cross of the F1 plants forms the F2 generation.
27
New cards
Mendel’s Hypotheses
1. Parents transmit information about a trait to their offspring in the form of what Mendel called “factors known as genes.
2. There are alternative versions of genes, called alleles. For example, purple and white are two alleles for the gene for flower color. The particular combination of alleles an organism has is called its genotype. The physical outcome that an organism displays as result of its genotype is called its phenotype.
3. For each inherited character, an organism inherits two alleles, one from each parent. An organism is homozygous for that gene if both alleles are identical. An organism is heterozygous for that gene if the alleles are different.
4. If two alleles of an inherited pair differ, then one determines the organism’s appearance and is called the dominant allele. The other has no noticeable effect on the organism’s appearance and is called the recessive allele. Dominant alleles are a capital letter; recessive alleles a lower case letters.
28
New cards
Inheritance many not follow Mendeilan genetics if:
.When alleles are not completely dominant or recessive
.When a gene has more than two alleles
.When a gene produces multiple phenotypes
.When a gene has more than two alleles
.When a gene produces multiple phenotypes
29
New cards
incomplete dominance
heterozygotes have an intermediate (blended) appearance between the phenotypes of the two alleles.
30
New cards
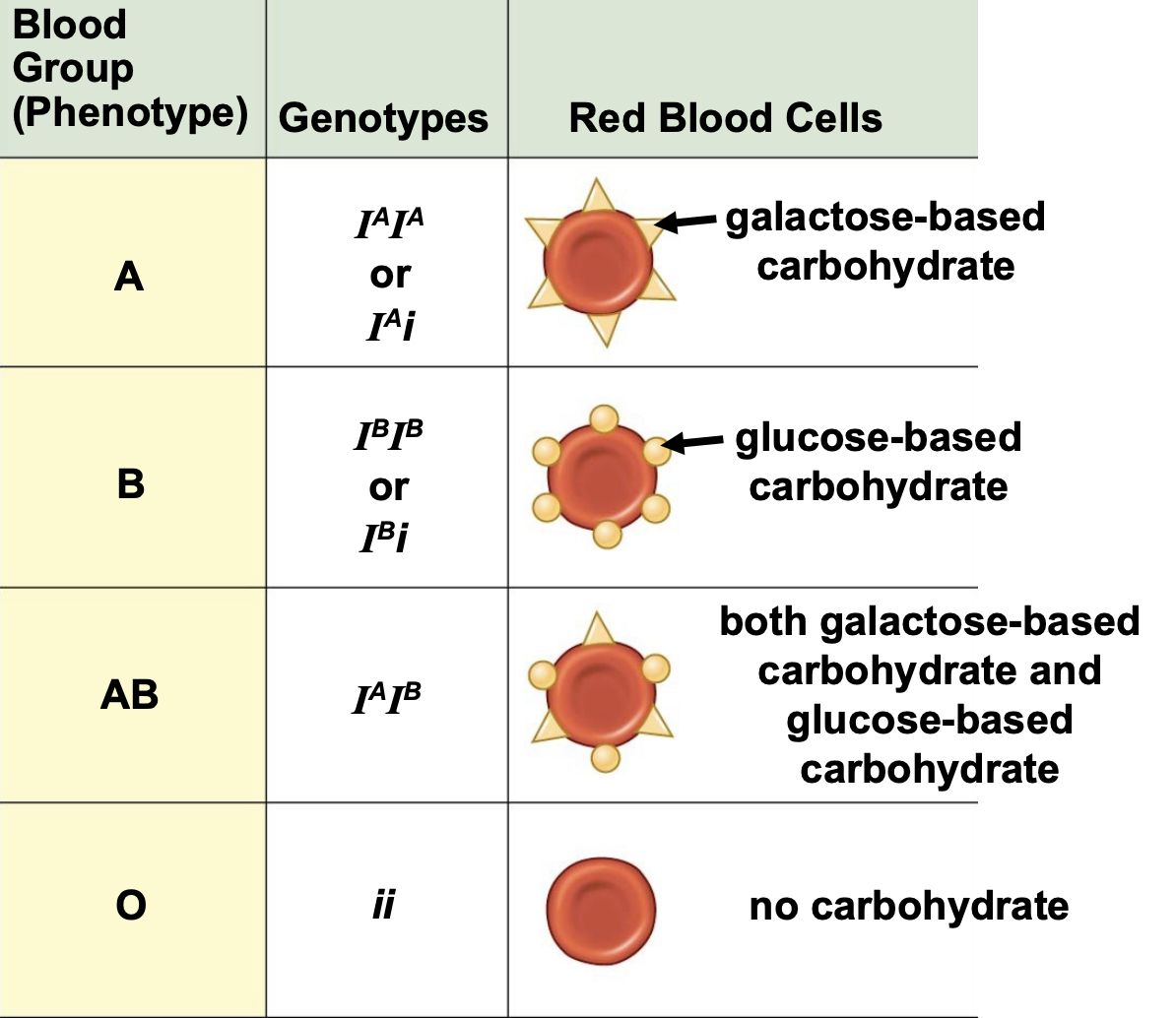
codominance and example
heterozygotes simultaneously display both alleles. They are denoted with capital letters and a superscript. ABO blood type, which exhibits both codominance and multiple alleles are example.
31
New cards
Blood Type example
Phenotype: A, B, AB, O
Genotype: I^A I^A, I^A i, I^B I^B, I^B i, I^A I^B, ii
Red Blood cells: Glucose, galactose, both, neither
Genotype: I^A I^A, I^A i, I^B I^B, I^B i, I^A I^B, ii
Red Blood cells: Glucose, galactose, both, neither
32
New cards
Nature and Nurture
The Environmental Impact on Phenotype. Traits that depend on multiple genes combined with environmental influences are called multifactorial
33
New cards
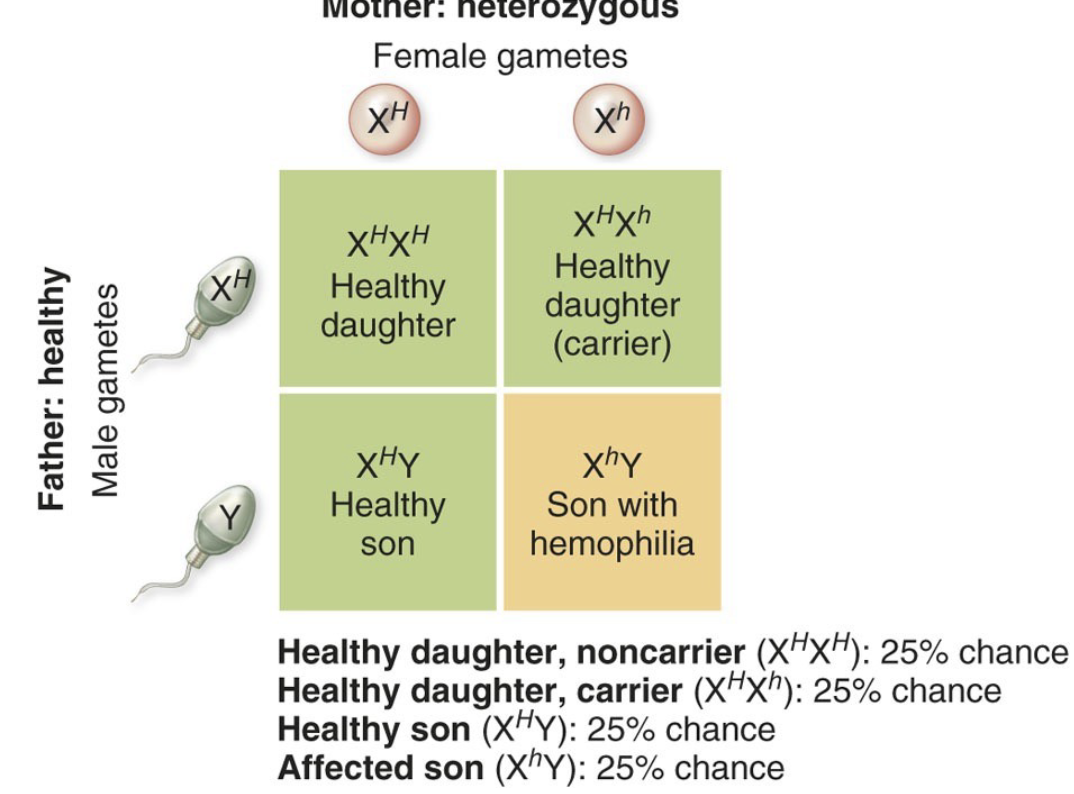
Sex-Linked Genes
Females must receive a recessive allele on both X chromosomes to express an X-linked recessive disorder. But a male only inherits one allele of an X-linked gene (because he gets a Y chromosome instead of a second X)
34
New cards
Pedigrees
**used to track genetic traits in a family**
35
New cards
law of independent assortment
the inheritance of one gene has no effect on the inheritance of another. An example is dihybrid cross where were crossing multiple alleys such as color and shape
36
New cards
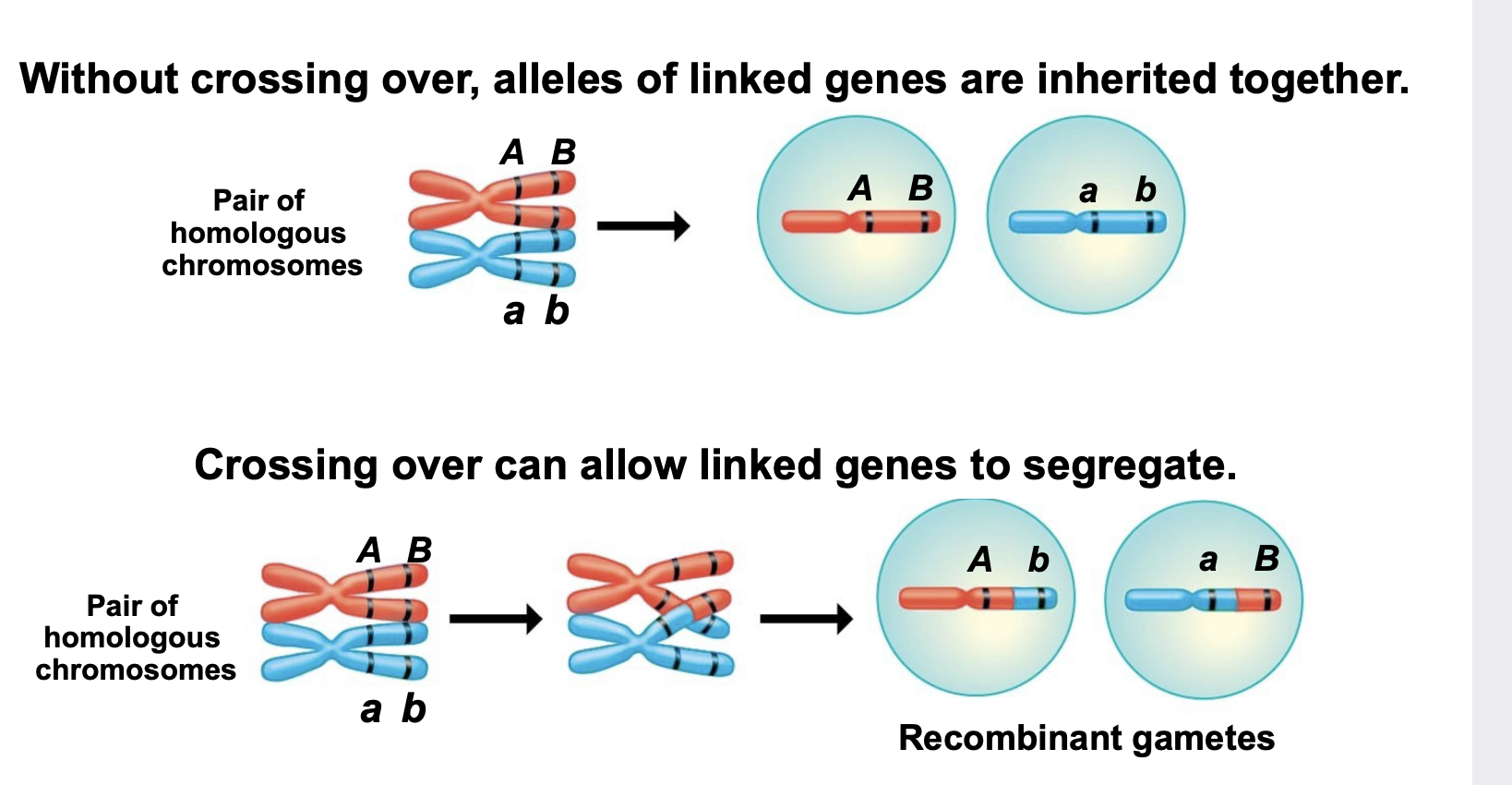
Linked genes and example
.located on the same chromosome and therefore tend to be inherited together and are thus exceptions to Mendel’s law of independent assortment
.Crossing over can allow occasional segregation of linked genes
.For example, the A and B genes are linked (located on the same chromosome). Therefore the alleles of the genes on the same chromosome will stay together (will not segregate) during gamete formation unless crossing over occurs to produce recombinants. Without crossing over, alleles of linked genes are inherited together. Crossing over can allow linked genes to segregate.
.Crossing over can allow occasional segregation of linked genes
.For example, the A and B genes are linked (located on the same chromosome). Therefore the alleles of the genes on the same chromosome will stay together (will not segregate) during gamete formation unless crossing over occurs to produce recombinants. Without crossing over, alleles of linked genes are inherited together. Crossing over can allow linked genes to segregate.
37
New cards
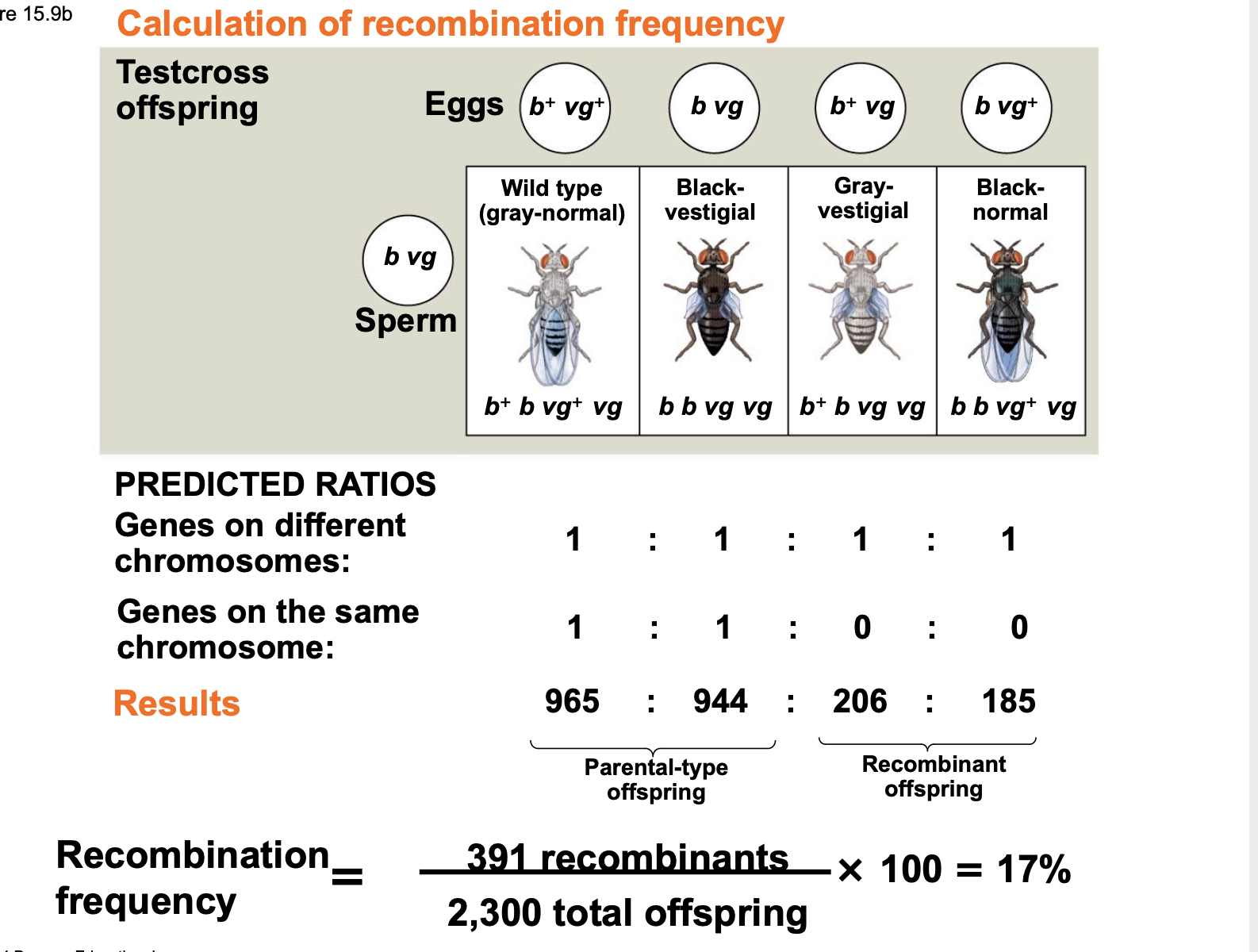
Recombination of Unlinked Genes: Independent Assortment of Chromosomes
.Offspring with a phenotype matching one of the parental phenotypes are called parental types.
.Offspring with nonparental phenotypes (new combinations of traits) are called recombinant types, or recombinants \n .A 50% frequency of recombination is observed for any two genes on different chromosomes (genes that are not linked) \n .Recombination frequencies of less than 50% indicate that genes are linked.
.Offspring with nonparental phenotypes (new combinations of traits) are called recombinant types, or recombinants \n .A 50% frequency of recombination is observed for any two genes on different chromosomes (genes that are not linked) \n .Recombination frequencies of less than 50% indicate that genes are linked.
38
New cards
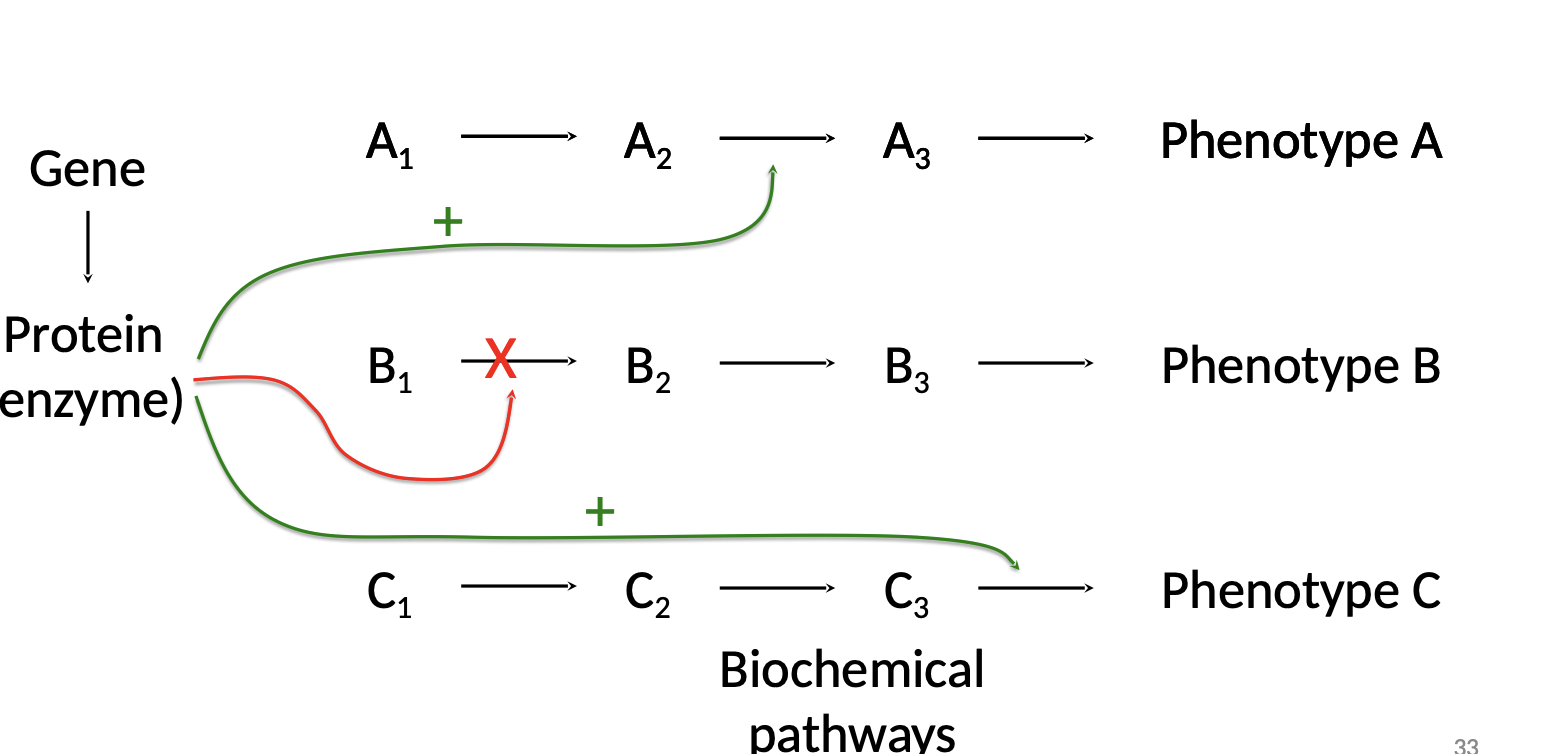
Pleiotropy
Pleiotropy is when one gene influences several characteristics. (One Gene, Many Phenotypes)
.Example: disease-causing genes
.Sickle-cell disease
\-results in abnormal hemoglobin proteins, and
\-causes disk-shaped red blood cells to deform into a sickle shape with jagged edges.
\-Sickled cells can lead to a cascade of symptoms, such as weakness, pain, organ damage, and paralysis.
.Example: disease-causing genes
.Sickle-cell disease
\-results in abnormal hemoglobin proteins, and
\-causes disk-shaped red blood cells to deform into a sickle shape with jagged edges.
\-Sickled cells can lead to a cascade of symptoms, such as weakness, pain, organ damage, and paralysis.
39
New cards
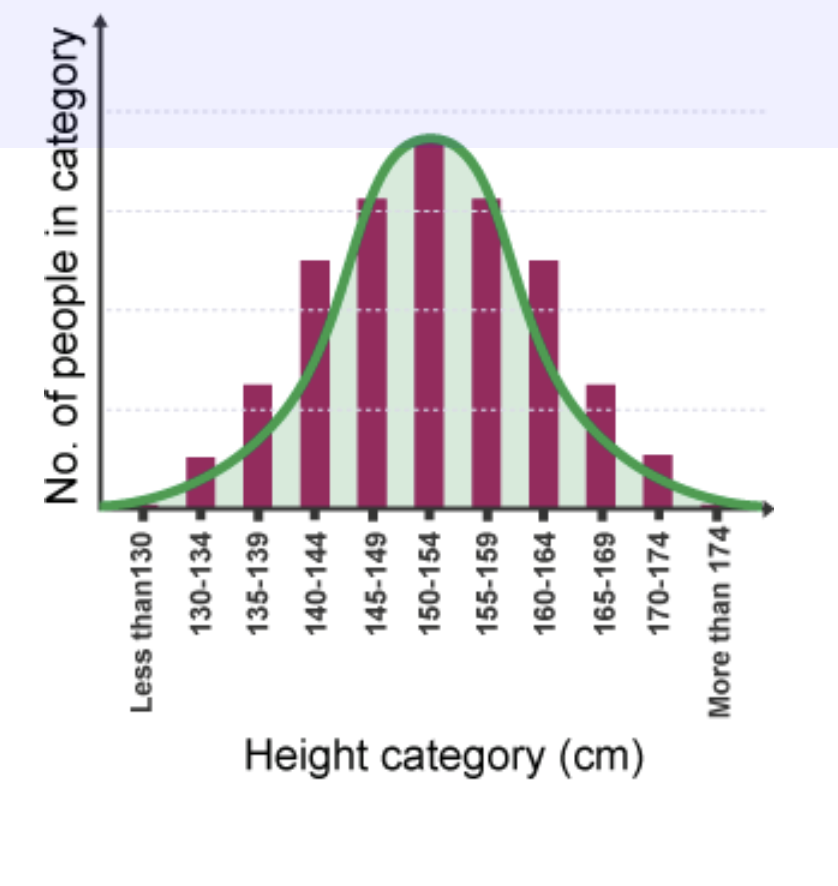
Polygenic Inheritance
the additive effects of two or more genes on a single phenotype.
.The more genes involved, and the more alleles of each of gene, the more phenotypes are possible.
.Examples: skin color, eye color, facial appearance, and height.
.Polygenic inheritance can also lead to continuous variation.
.Characters can show a range of small differences when multiple genes act jointly to influence a phenotype.
The gradation in phenotypes is called continuous variation. A good example is human height.
.The more genes involved, and the more alleles of each of gene, the more phenotypes are possible.
.Examples: skin color, eye color, facial appearance, and height.
.Polygenic inheritance can also lead to continuous variation.
.Characters can show a range of small differences when multiple genes act jointly to influence a phenotype.
The gradation in phenotypes is called continuous variation. A good example is human height.
40
New cards
Epistasis
.expression of a gene at one locus alters the phenotypic expression of a gene at a second locus.
.For example, in Labrador retrievers and many other mammals, coat color depends on two genes.
• One gene determines the pigment color (with alleles B for black and b for brown).
• The other gene (with alleles E for color and e for no color) determines whether the pigment will be deposited in the hair.
.For example, in Labrador retrievers and many other mammals, coat color depends on two genes.
• One gene determines the pigment color (with alleles B for black and b for brown).
• The other gene (with alleles E for color and e for no color) determines whether the pigment will be deposited in the hair.
41
New cards
Genomic Imprinting
.For a few mammalian traits, the phenotype depends on which parent passed along the alleles for those traits.
.Such variation in phenotype is called genomic imprinting.
.Genomic imprinting involves the silencing of certain genes depending on which parent passes them on.
.imprinting is the result of the methylation (addition of —CH3) of cysteine nucleotides in the DNA of one parent but not the other.
.Methylation inhibits expression of the modified genes.
.Genomic imprinting is thought to affect only a small fraction of mammalian genes.
.Most imprinted genes are critical for embryonic development
.Such variation in phenotype is called genomic imprinting.
.Genomic imprinting involves the silencing of certain genes depending on which parent passes them on.
.imprinting is the result of the methylation (addition of —CH3) of cysteine nucleotides in the DNA of one parent but not the other.
.Methylation inhibits expression of the modified genes.
.Genomic imprinting is thought to affect only a small fraction of mammalian genes.
.Most imprinted genes are critical for embryonic development
42
New cards
Inheritance of Organelle Genes
.Extranuclear genes (or cytoplasmic genes) are found in organelles in the cytoplasm.
.Mitochondria, chloroplasts, and other plant plastids carry small circular DNA molecules.
.Extranuclear genes are inherited maternally because the zygote’s cytoplasm comes from the egg
.Mitochondria, chloroplasts, and other plant plastids carry small circular DNA molecules.
.Extranuclear genes are inherited maternally because the zygote’s cytoplasm comes from the egg
43
New cards
nucleotides
monomers of nucleic acids, which are polymers. Each nucleotide consists of a nitrogenous base, a pentose sugar, and a phosphate group. The portion of a nucleotide without the phosphate group (sugar + base) is called a nucleoside. There are two types of nucleic acids, DNA (deoxyribonucleic acid) and RNA (ribonucleic acid), which differ in the sugars and bases that make up their respective nucleotides.
44
New cards
two families of nitrogenous bases
.Pyrimidines (cytosine, thymine, and uracil) have a single six-membered ring.
.Purines (adenine and guanine) have a double ring (a six-membered ring fused to a five-membered ring)
.Purines (adenine and guanine) have a double ring (a six-membered ring fused to a five-membered ring)
45
New cards
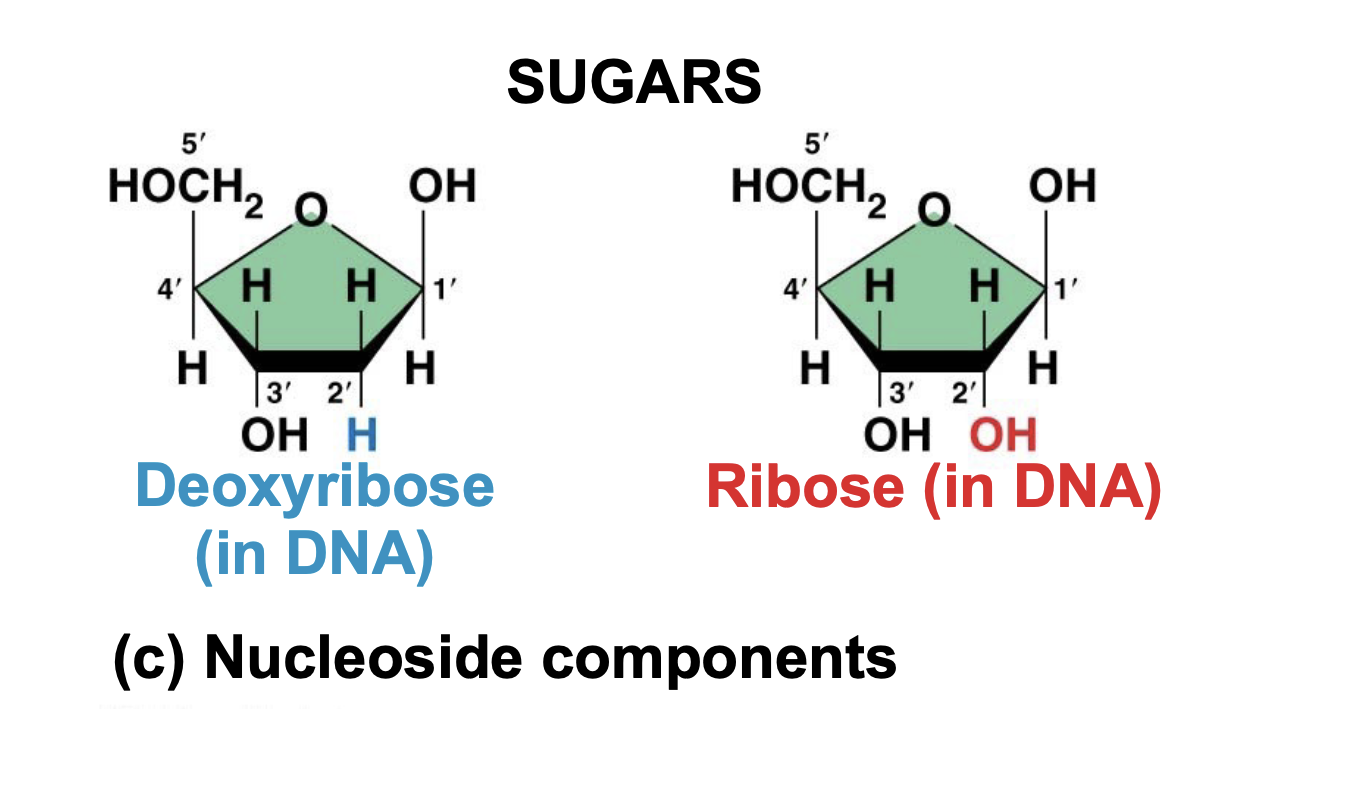
DNA and RNA difference
.DNA uses thymine, whereas RNA uses uracil instead.
. DNA and RNA also differ in the sugar used in their nucleotides.
. DNA and RNA also differ in the sugar used in their nucleotides.
46
New cards
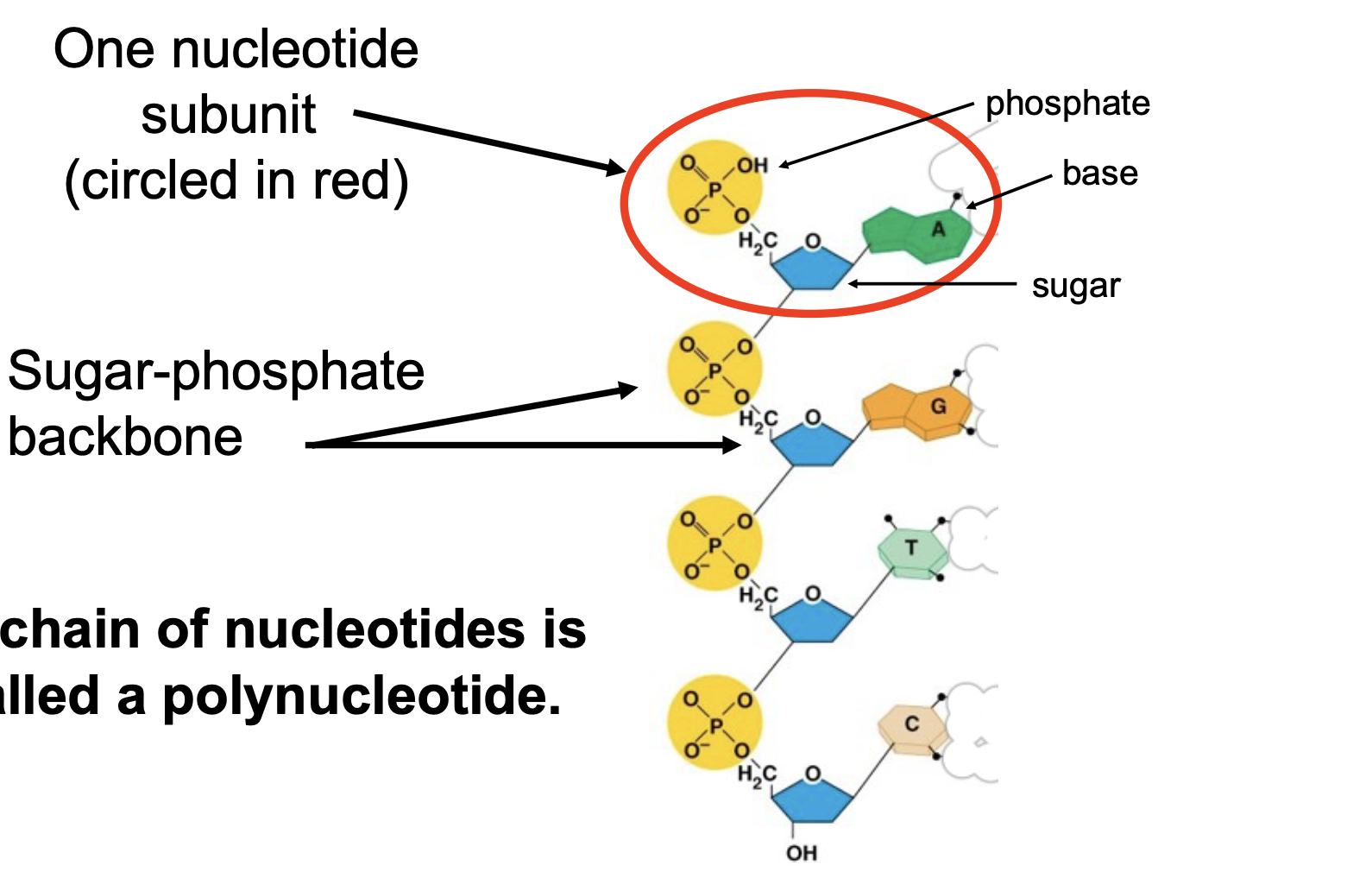
How is DNA polymer formed
the phosphate group of one nucleotide is covalently bonded to the sugar of the next. This forms what is called the “sugar phosphate backbone” of DNA.
47
New cards
polynucleotide
A chain of nucleotides. The two polynucleotide chains in a double helix are antiparallel
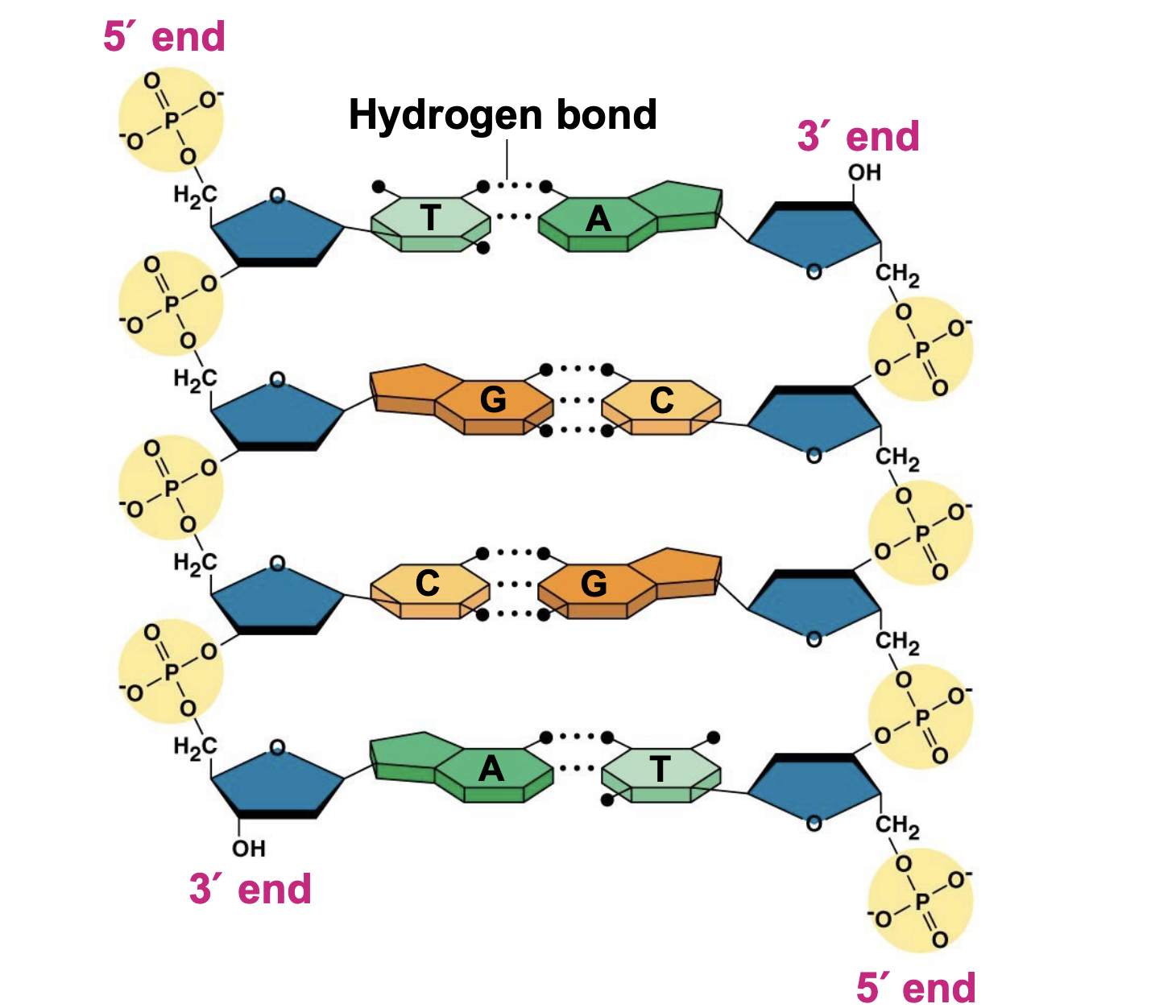
48
New cards
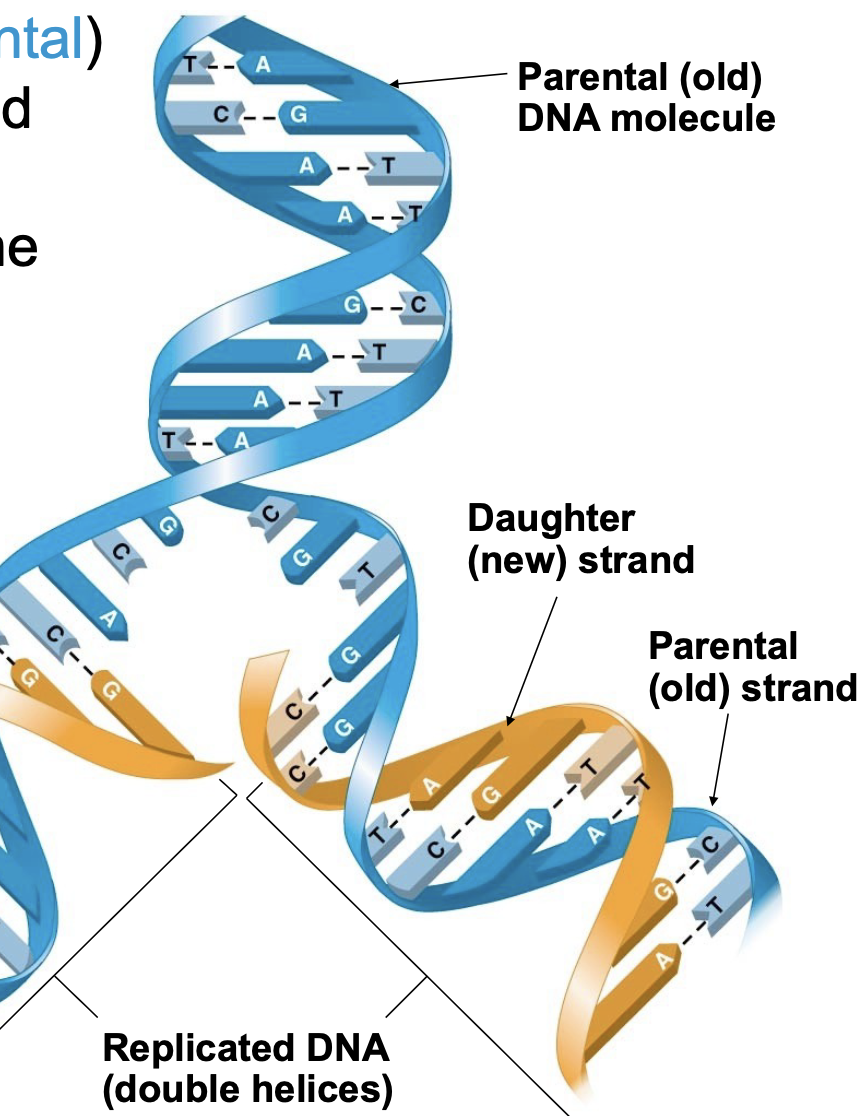
What happens in DNA replications
.the parent molecule unwinds, and two new daughter strands are built based on base- pairing rules. Strands of the original (parental) DNA helix are separated, and new (daughter) strands complementary to each of the parental strands are made. Thus, each duplicated DNA \\n double helix contains one old strand and one new strand. This is called semi-conservative replication.
49
New cards
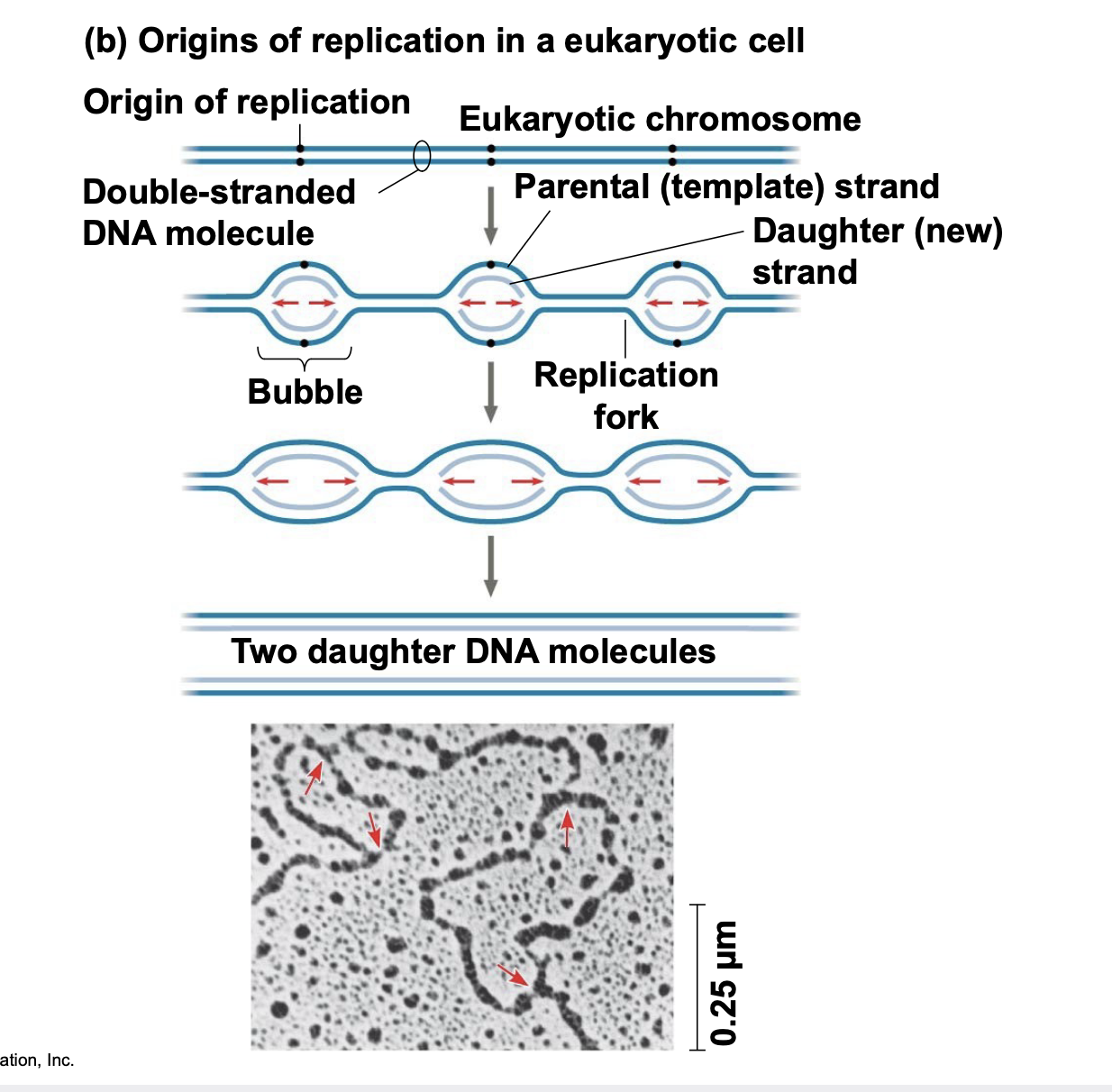
origins of replications
.Replication begins at particular sites called origins of replication, where the two DNA strands are separated, opening up a replication “bubble”
.A eukaryotic chromosome may have hundreds or even thousands of origins of replication
.Replication proceeds in both directions from each origin, until the entire molecule is copied
.A eukaryotic chromosome may have hundreds or even thousands of origins of replication
.Replication proceeds in both directions from each origin, until the entire molecule is copied
50
New cards
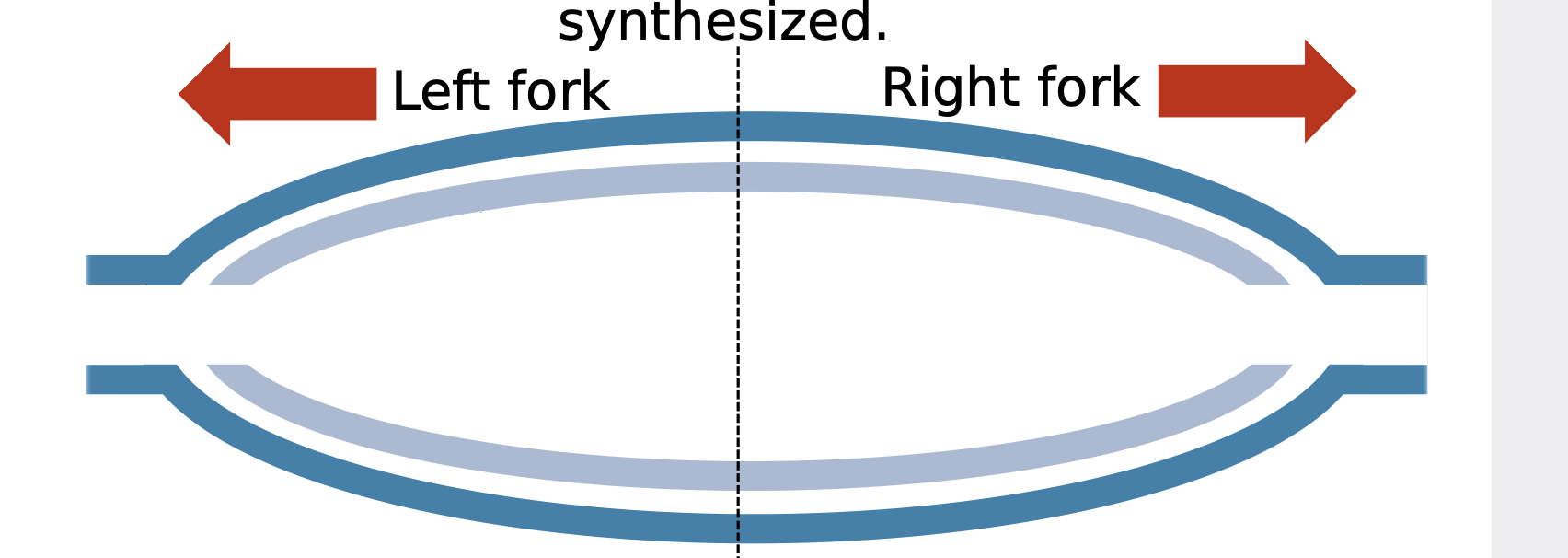
How is replication started
.A replication bubble is divided into two replication forks, where the two strands of the original double-stranded DNA (dark blue) are pulled apart and new DNA (light blue/grey) is synthesized. The bubble will widen as the two forks extend in opposite directions (red arrows) as more of the original DNA is unwound and replicated.
51
New cards
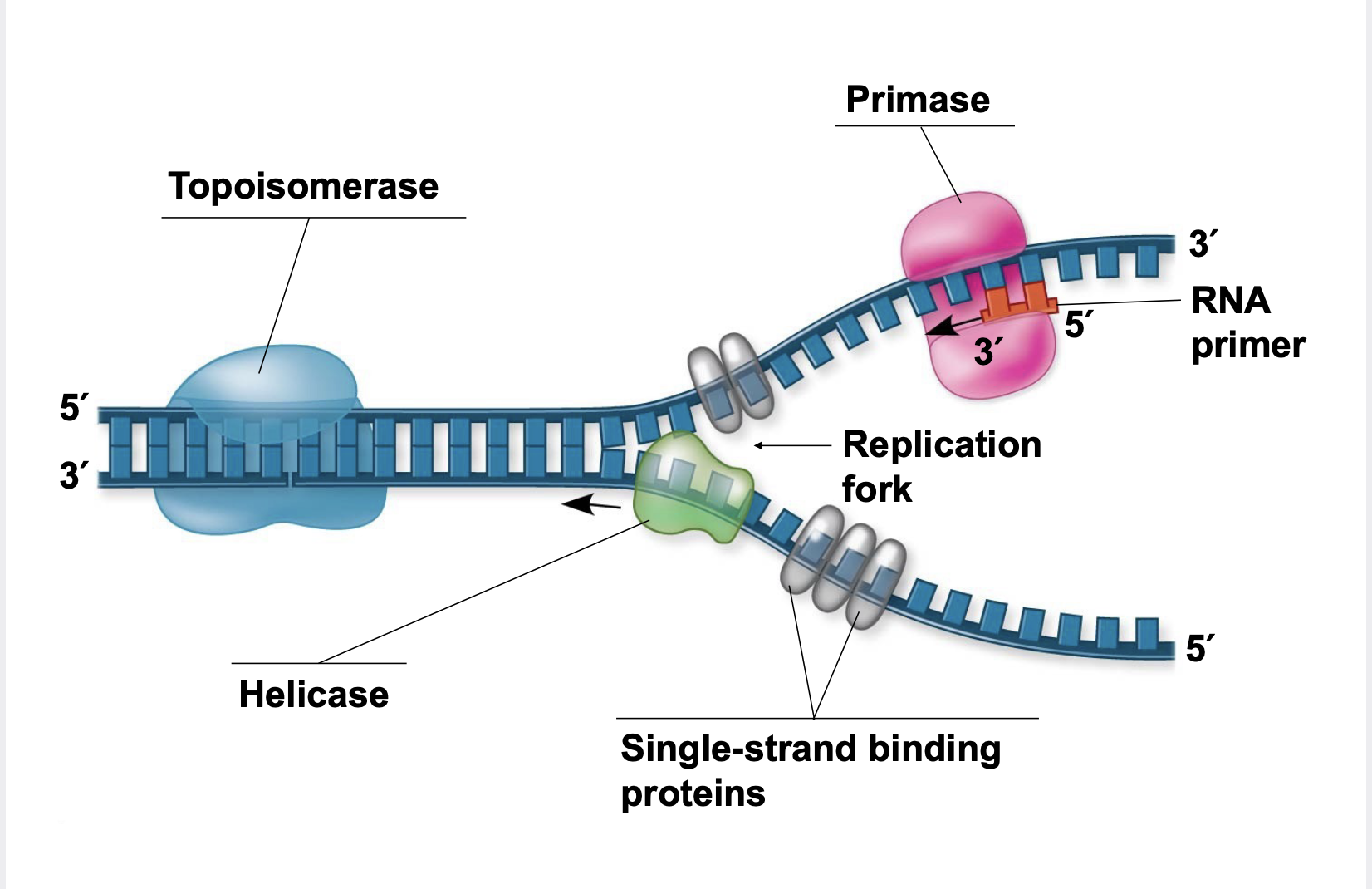
replication fork, Helicases, Single-strand binding proteins, Topoisomerase
At the end of each replication bubble is a replication fork, a Y-shaped region where new DNA strands are elongating.
.Helicases are enzymes that untwist the double helix at the replication forks
.Single-strand binding proteins bind to and stabilize single-stranded DNA
.Topoisomerase corrects “overwinding” ahead of replication forks by breaking, swiveling, and rejoining DNA strands
.DNA polymerases cannot initiate synthesis of a polynucleotide; they can only add nucleotides to an existing 3′ end \n .The initial nucleotide strand is a short RNA primer \n .An enzyme called primase can start an RNA chain from scratch and adds RNA nucleotides one at a time using the parental DNA as a template \n .The primer is short (5–10 nucleotides long), and the 3′ end serves as the starting point for the new DNA strand
.Helicases are enzymes that untwist the double helix at the replication forks
.Single-strand binding proteins bind to and stabilize single-stranded DNA
.Topoisomerase corrects “overwinding” ahead of replication forks by breaking, swiveling, and rejoining DNA strands
.DNA polymerases cannot initiate synthesis of a polynucleotide; they can only add nucleotides to an existing 3′ end \n .The initial nucleotide strand is a short RNA primer \n .An enzyme called primase can start an RNA chain from scratch and adds RNA nucleotides one at a time using the parental DNA as a template \n .The primer is short (5–10 nucleotides long), and the 3′ end serves as the starting point for the new DNA strand
52
New cards
Synthesizing a New DNA Strand
.Enzymes called DNA polymerases synthesize new DNA \n .DNA polymerases require a primer (another nucleic acid to add new nucleotides to) and a DNA template strand. \n .DNA polymerases can add nucleotides only to the 3 ́ end of a primer. \n .DNA polymerases use deoxynucleoside triphosphates, but only one of the three phosphates is added into the DNA chain.
.Each nucleotide that is added to a growing DNA strand is a deoxynucleoside triphosphate.
.As each monomer (in triphosphate form) joins the DNA strand, it loses two phosphate groups, and thus only a single phosphate is placed into the sugar- phosphate backbone.
.Each nucleotide that is added to a growing DNA strand is a deoxynucleoside triphosphate.
.As each monomer (in triphosphate form) joins the DNA strand, it loses two phosphate groups, and thus only a single phosphate is placed into the sugar- phosphate backbone.
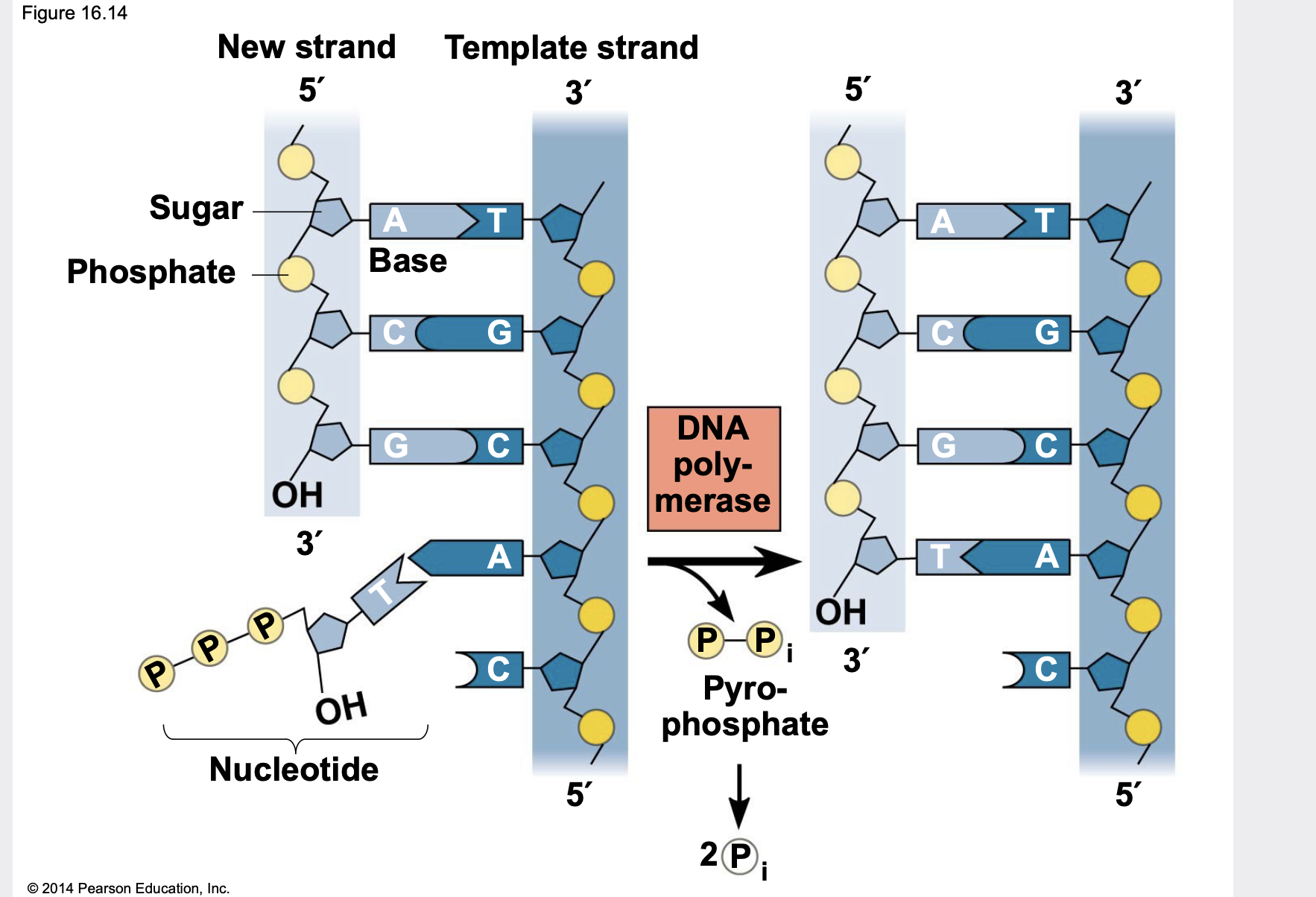
53
New cards
Antiparallel Elongation
.The antiparallel structure of the double helix affects replication \\n .DNA polymerases add nucleotides only to the free 3′ end of a growing strand; therefore, a new DNA strand can elongate only in the 5′ to 3′ direction
.DNA polymerase synthesizes a leading strand continuously, moving toward the replication fork.
To elongate the other new strand, called the lagging strand, DNA polymerase must work in the direction away from the replication fork (to add nucleotides 5 ́ \\n to 3 ́)
.The lagging strand is synthesized as a series of segments called Okazaki fragments, which are joined together by DNA ligase
.As the replication fork moves leftward, one new strand (called the leading strand) can be polymerized in its 5 ́ to 3 ́ direction without stopping. The other new strand (called the lagging strand) must be made by repeatedly re-starting polymerization because its 5 ́ to 3 ́ direction is opposite the replication fork direction. Leading strand DNA synthesis extends 5 ́ to 3 ́ in the same direction as the fork. Lagging strand DNA synthesis extends 5 ́ to 3 ́ in the opposite direction as the fork.
.DNA polymerase synthesizes a leading strand continuously, moving toward the replication fork.
To elongate the other new strand, called the lagging strand, DNA polymerase must work in the direction away from the replication fork (to add nucleotides 5 ́ \\n to 3 ́)
.The lagging strand is synthesized as a series of segments called Okazaki fragments, which are joined together by DNA ligase
.As the replication fork moves leftward, one new strand (called the leading strand) can be polymerized in its 5 ́ to 3 ́ direction without stopping. The other new strand (called the lagging strand) must be made by repeatedly re-starting polymerization because its 5 ́ to 3 ́ direction is opposite the replication fork direction. Leading strand DNA synthesis extends 5 ́ to 3 ́ in the same direction as the fork. Lagging strand DNA synthesis extends 5 ́ to 3 ́ in the opposite direction as the fork.
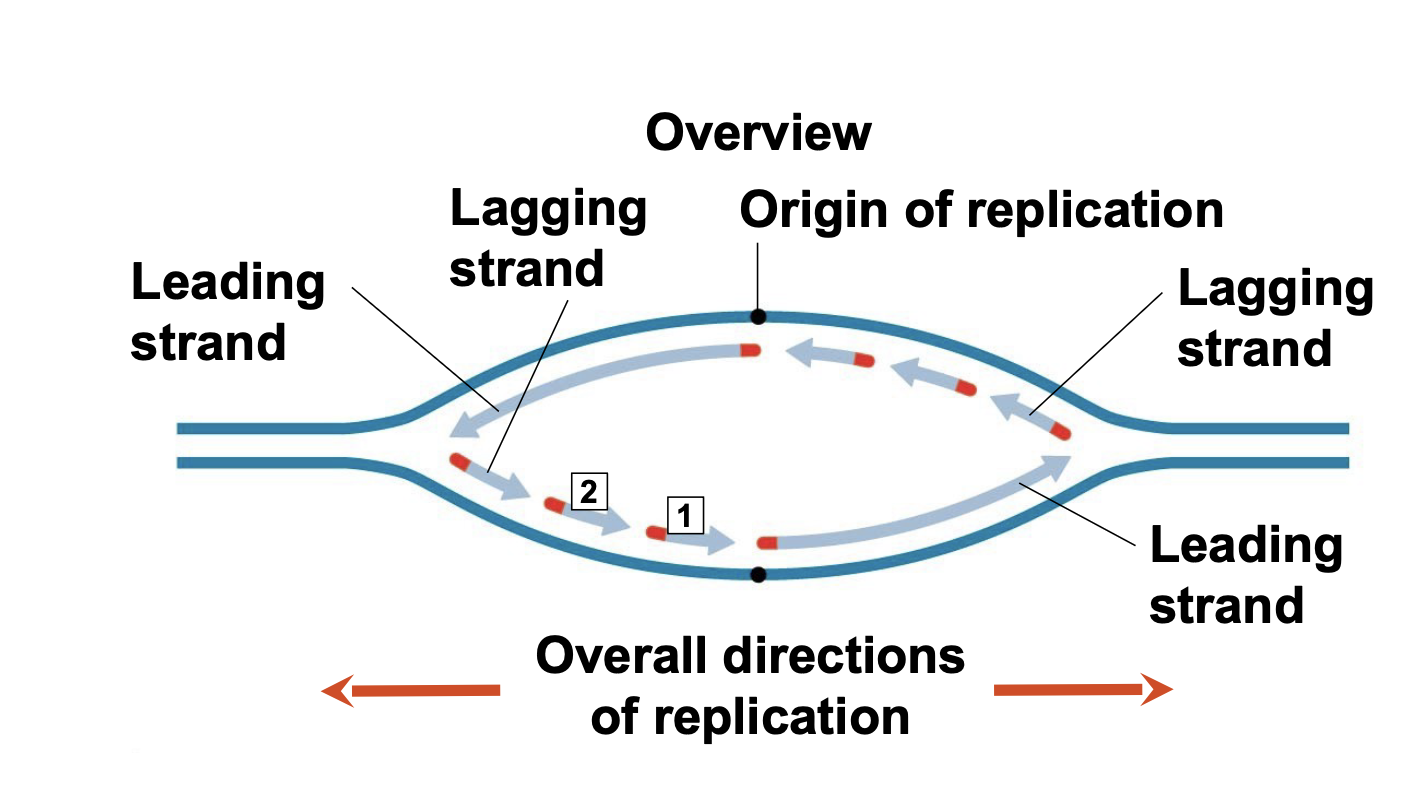
54
New cards
Lagging strand synthesis steps
1. Primase makes RNA primer.
2. DNA pol III makes Okazaki fragment 1
3. DNA pol III detaches.
4. DNA pol III makes Okazaki fragment 2.
5. DNA pol I replaces RNA with DNA
6. DNA ligase forms bonds between DNA fragments.
55
New cards
Types of DNA polymerases:
DNA Polymerase III (Pol III) polymerizes long stretches of nucleotides during replication. \n DNA Polymerase I (Pol I) replaces the primer RNA with DNA during replication.
56
New cards
The DNA Replication Complex (Replisome)
.The proteins that participate in DNA replication form a large complex called the replisome.
The replisome is stationary during the replication process. DNA polymerase molecules “reel in” parental DNA and “extrude” newly made daughter DNA molecules.
The replisome is stationary during the replication process. DNA polymerase molecules “reel in” parental DNA and “extrude” newly made daughter DNA molecules.
57
New cards
Gene expression in DNA and proteins
DNA stores information in the sequence of nucleotides.
.The DNA inherited by an organism leads to specific traits by dictating the synthesis of proteins.
.Proteins are the links between genotype and phenotype \n .Gene expression is the process in which DNA directs protein synthesis. This includes two basic stages: transcription and translation
.The DNA inherited by an organism leads to specific traits by dictating the synthesis of proteins.
.Proteins are the links between genotype and phenotype \n .Gene expression is the process in which DNA directs protein synthesis. This includes two basic stages: transcription and translation
58
New cards
Basic Principles of Transcription and Translation
RNA is the bridge between genes and the proteins for which they code
.Transcription is the synthesis of RNA using information in DNA.
.Transcription produces RNAs such as messenger RNA (mRNA), which is an RNA copy of a gene.
.Translation is the synthesis of a polypeptide (protein) using information in the mRNA.
.Ribosomes are the enzymes that perform translation
.Transcription is the synthesis of RNA using information in DNA.
.Transcription produces RNAs such as messenger RNA (mRNA), which is an RNA copy of a gene.
.Translation is the synthesis of a polypeptide (protein) using information in the mRNA.
.Ribosomes are the enzymes that perform translation
59
New cards
Language in transcription and translations
Language is a recording of information.
.The information is initially recorded in the language of nucleic acids, written in DNA nucleotides.
.The information is transcribed (re-written) into mRNA, still in the nucleic acid language, but now written in RNA nucleotides.
.Finally, the information is translated (changed from one language to another) into protein language written in amino acids.
.The information is initially recorded in the language of nucleic acids, written in DNA nucleotides.
.The information is transcribed (re-written) into mRNA, still in the nucleic acid language, but now written in RNA nucleotides.
.Finally, the information is translated (changed from one language to another) into protein language written in amino acids.
60
New cards
RNA polymerase
RNA synthesis is catalyzed by RNA polymerase.
.RNA polymerase separates the DNA strands then uses one of the strands as a template to polymerize nucleotides into RNA.
.The RNA sequence is complementary to the sequence of DNA template strand
\-RNA polymerase does not need any primer
\-RNA synthesis follows the same base-pairing rules as DNA, except that uracil substitutes for thymine
.RNA polymerase separates the DNA strands then uses one of the strands as a template to polymerize nucleotides into RNA.
.The RNA sequence is complementary to the sequence of DNA template strand
\-RNA polymerase does not need any primer
\-RNA synthesis follows the same base-pairing rules as DNA, except that uracil substitutes for thymine
61
New cards
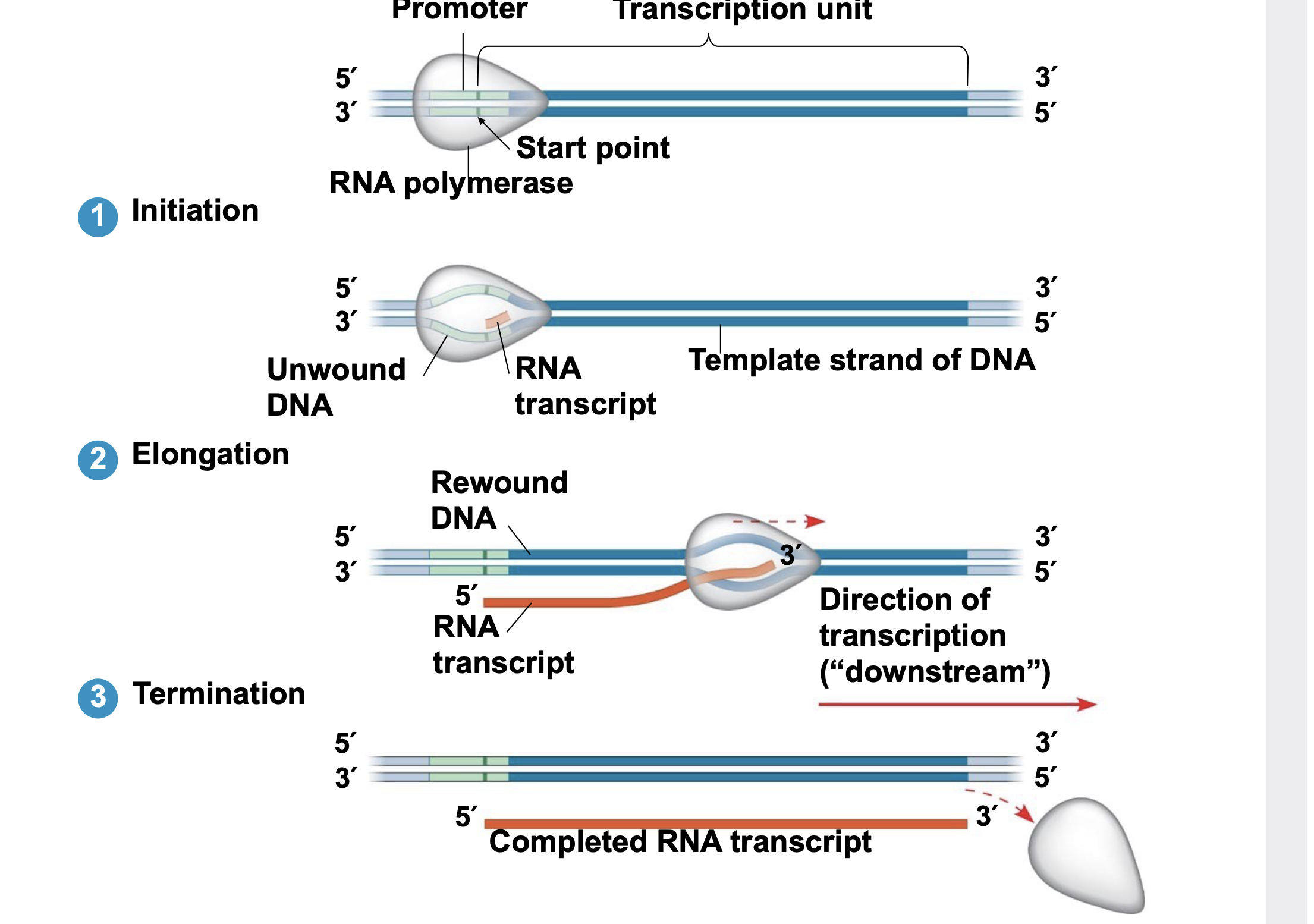
promoter, terminator, transcription unit
The DNA sequence where RNA polymerase binds the DNA to begin transcription is called the promoter.
In bacteria, the sequence signaling the end of transcription is called the terminator. Eukaryotes do not use a terminator sequence.
.Transcription does not copy the entire chromosome into RNA; it only copies a gene at a time. The sequence of DNA that is transcribed is called a transcription unit.
In bacteria, the sequence signaling the end of transcription is called the terminator. Eukaryotes do not use a terminator sequence.
.Transcription does not copy the entire chromosome into RNA; it only copies a gene at a time. The sequence of DNA that is transcribed is called a transcription unit.
62
New cards
The three stages of transcription
1. initiation
2. elongation
3. termination
63
New cards
Initiation
.Promoters mark the transcriptional start point and usually extend several dozen nucleotide pairs upstream of the start point.
.Transcription factors are proteins that bind to the promoter and are required for the binding of RNA polymerase and the initiation of transcription.
.The completed assembly of transcription factors and RNA polymerase II bound to a promoter is called a transcription initiation complex.
.A sequence in the promoter called a TATA box is crucial in forming the initiation complex in eukaryotes
.Transcription factors are proteins that bind to the promoter and are required for the binding of RNA polymerase and the initiation of transcription.
.The completed assembly of transcription factors and RNA polymerase II bound to a promoter is called a transcription initiation complex.
.A sequence in the promoter called a TATA box is crucial in forming the initiation complex in eukaryotes
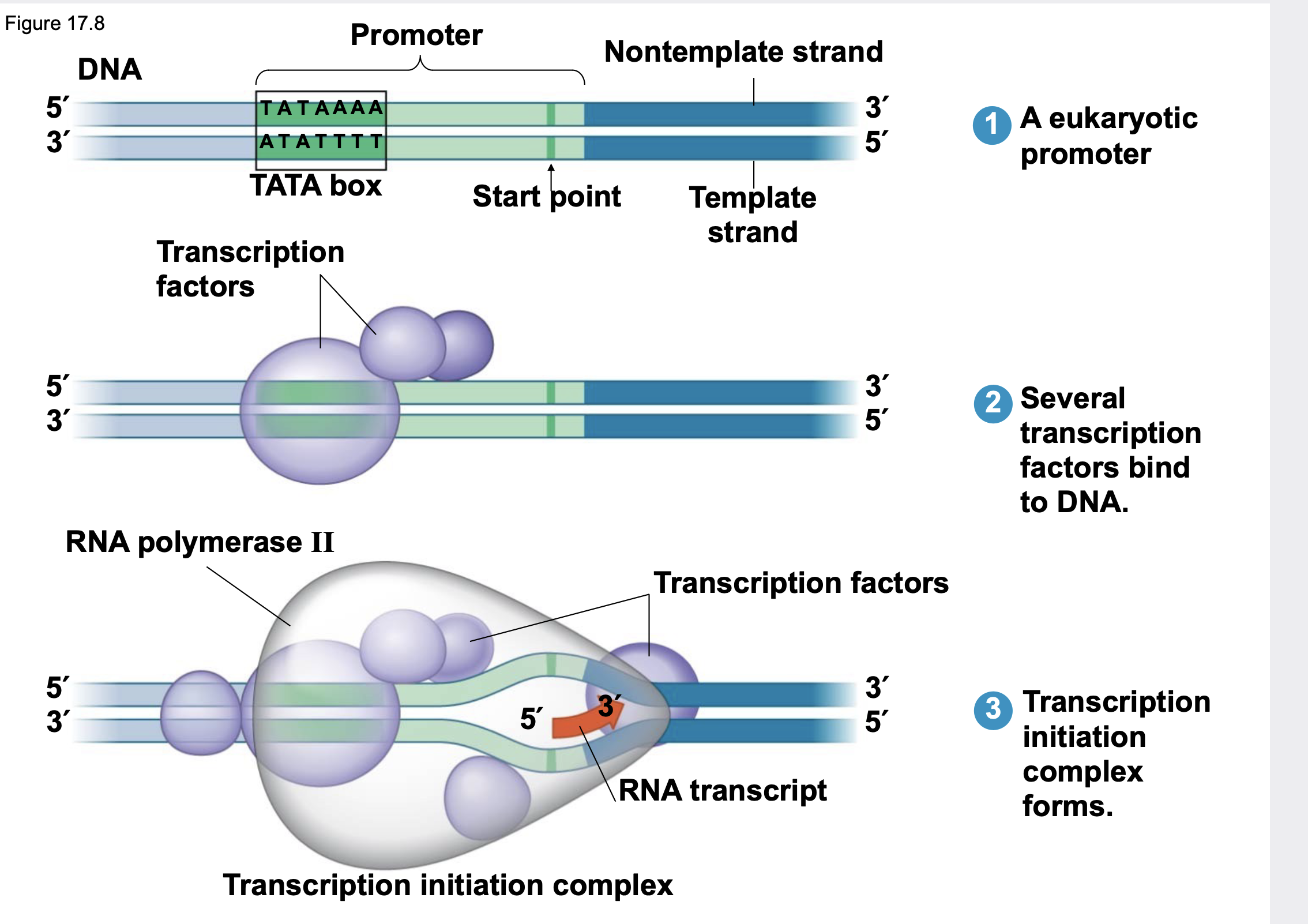
64
New cards
Elongation
.As RNA polymerase moves along the DNA, it unwinds the double helix, 10 to 20 bases at a time.
.Transcription progresses at a rate of 40 nucleotides per second in eukaryotes.
.A gene can be transcribed simultaneously by several RNA polymerases to make multiple RNAs.
.Nucleotides are added to the 3′ end of the growing RNA molecule according to complementary base pairing with the template strand of DNA.
RNA polymerase moves along the DNA, making an RNA copy in the 5 ́ to 3 ́ direction complementary to the DNA template sequence.
.Transcription progresses at a rate of 40 nucleotides per second in eukaryotes.
.A gene can be transcribed simultaneously by several RNA polymerases to make multiple RNAs.
.Nucleotides are added to the 3′ end of the growing RNA molecule according to complementary base pairing with the template strand of DNA.
RNA polymerase moves along the DNA, making an RNA copy in the 5 ́ to 3 ́ direction complementary to the DNA template sequence.
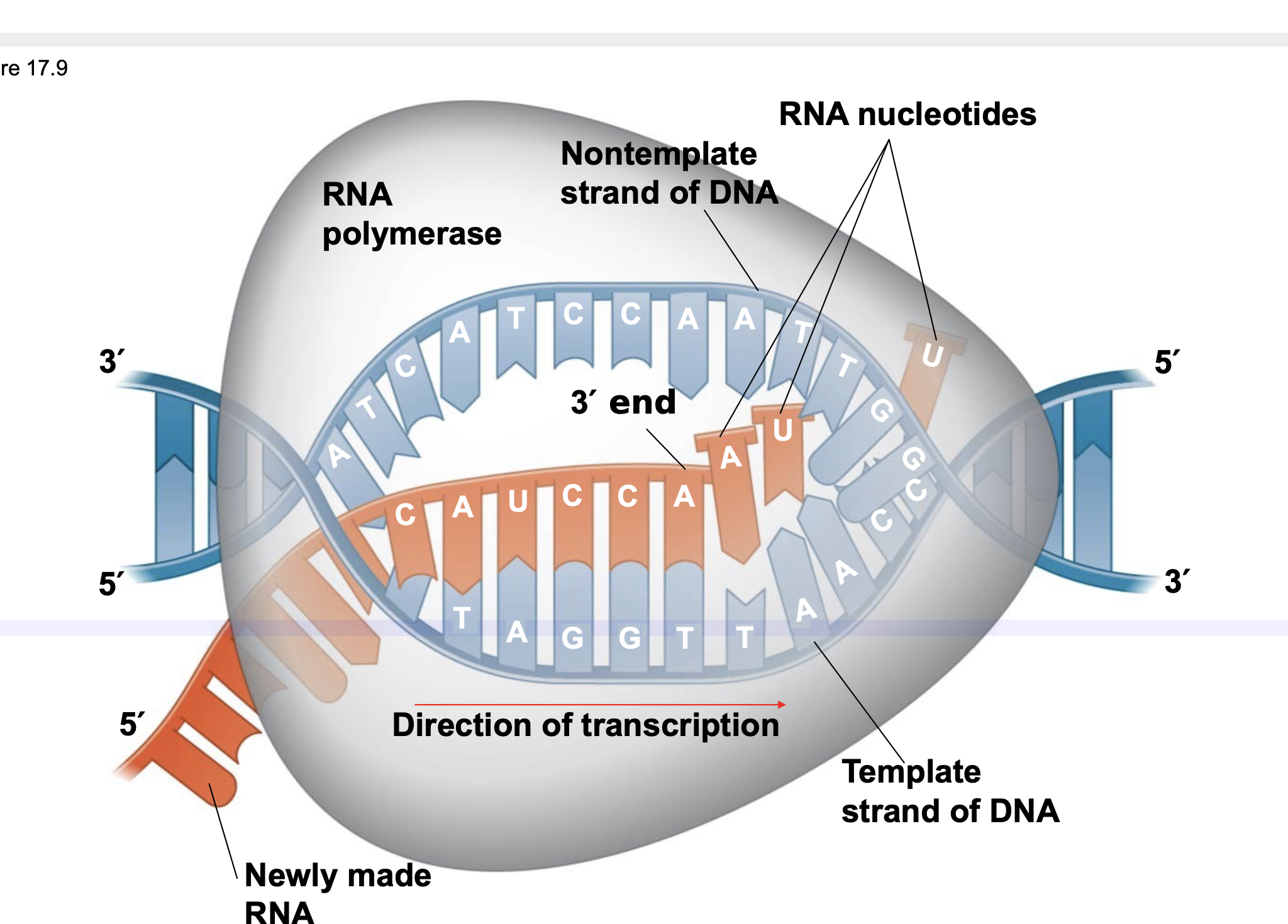
65
New cards
Termination
.The mechanisms of termination are different in bacteria and eukaryotes.
In bacteria, the polymerase stops transcription at the end of a specific sequence in the DNA called the terminator, and the mRNA can be translated without \\n further modification.
.In eukaryotes, RNA polymerase transcribes the polyadenylation signal sequence present at the end of a eukaryotic gene. The RNA transcript is released after the polymerase has transcribed 10–35 nucleotides past this polyadenylation sequence.
In bacteria, the polymerase stops transcription at the end of a specific sequence in the DNA called the terminator, and the mRNA can be translated without \\n further modification.
.In eukaryotes, RNA polymerase transcribes the polyadenylation signal sequence present at the end of a eukaryotic gene. The RNA transcript is released after the polymerase has transcribed 10–35 nucleotides past this polyadenylation sequence.
66
New cards
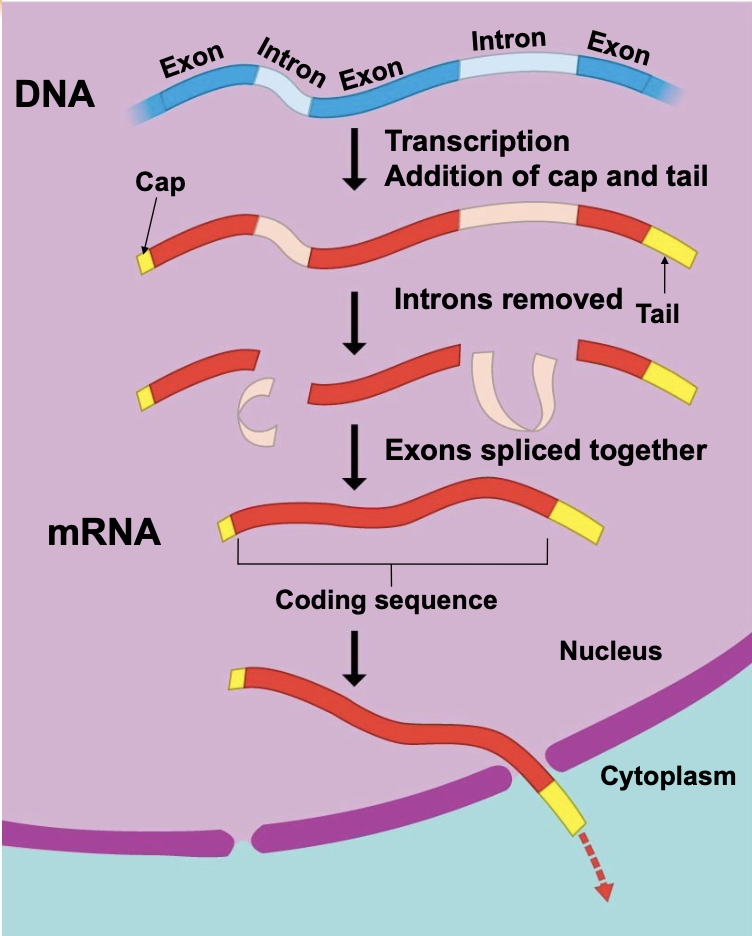
Eukaryotic cells modify RNA after transcription
Enzymes in the eukaryotic nucleus modify RNA (RNA processing) before they are sent to the cytoplasm. Messenger RNAs are called pre-mRNAs before they have been processed. \n ▪ During RNA processing in eukaryotes: \n 1. A “cap” (modified nucleotide) is added to the 5 ́ end of the pre-mRNA \n 2. A stretch of extra A nucleotides is added to the 3 ́ end (poly-A tail) \n 3. Sequences in the gene that do not encode protein are removed (splicing)
67
New cards
Alteration of mRNA Ends: 5 ́ cap and poly-A tail
Each end of a pre-mRNA molecule is modified in a particular way \n ▪ The 5′ end receives a modified nucleotide (7mG) called the 5′ cap \n ▪ The 3′ end gets a poly-A tail when an enzyme called poly A polymerase adds a bunch (50-250) of A nucleotides that are not encoded in the gene. \n ▪ These modifications share several functions \n ▪ They seem to help the export of mRNA to the cytoplasm \n ▪ They protect mRNA from hydrolytic enzymes (keep it from being chewed up by the cell) \n ▪ They help ribosomes bind the mRNA to begin translatio
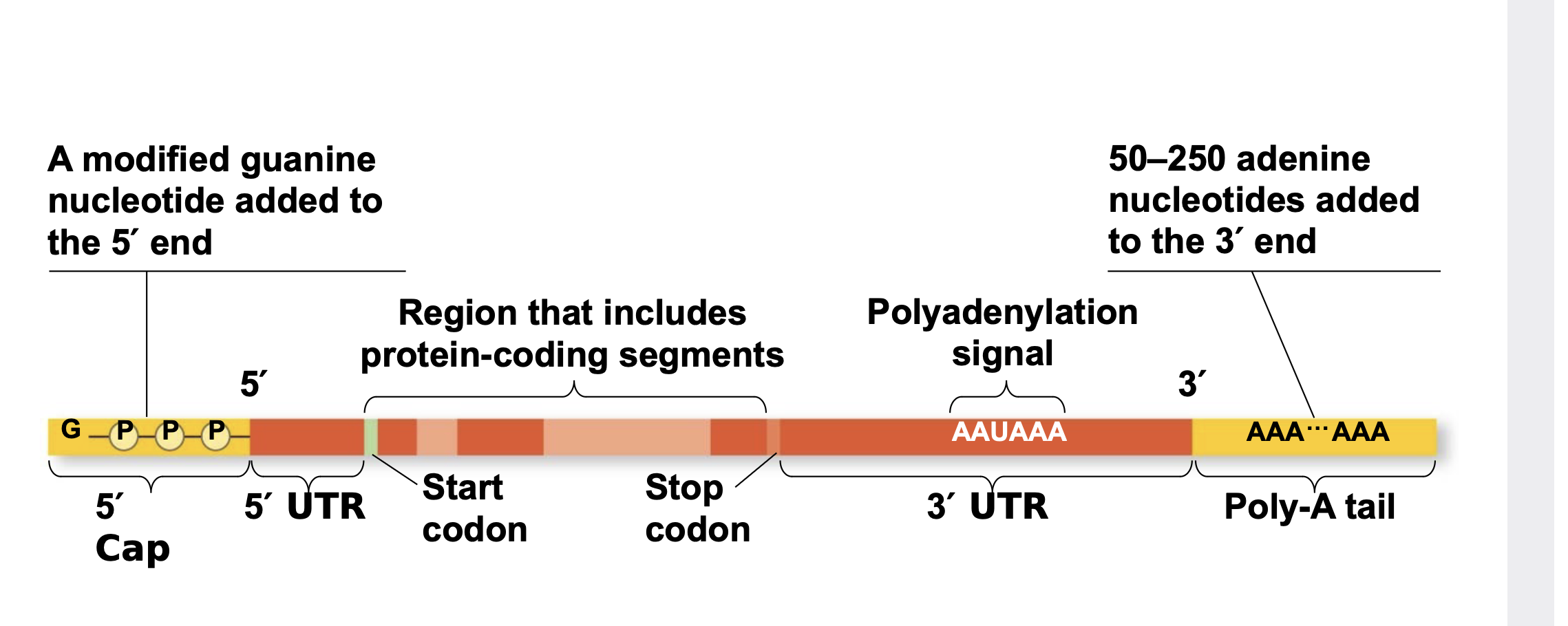
68
New cards
pre-mRNA Splicing
\
The function of mRNA is to store the nucleotide sequence that encodes proteins. However, most eukaryotic genes and their RNA transcripts have stretches of nucleotide sequence that do NOT encode protein and must be removed before the mRNA can be translated. \n ▪ These noncoding sequences are called introns. \n ▪ The sequences in a gene that do encode protein are called exons. \n ▪ pre-mRNA splicing removes the introns from an RNA and joins together the exons, creating an mRNA molecule with a continuous coding sequence that is ready for translation.
.In eukaryotes, the protein-coding exons are interrupted by non-coding introns which are removed from the pre-mRNA by splicing.
The function of mRNA is to store the nucleotide sequence that encodes proteins. However, most eukaryotic genes and their RNA transcripts have stretches of nucleotide sequence that do NOT encode protein and must be removed before the mRNA can be translated. \n ▪ These noncoding sequences are called introns. \n ▪ The sequences in a gene that do encode protein are called exons. \n ▪ pre-mRNA splicing removes the introns from an RNA and joins together the exons, creating an mRNA molecule with a continuous coding sequence that is ready for translation.
.In eukaryotes, the protein-coding exons are interrupted by non-coding introns which are removed from the pre-mRNA by splicing.
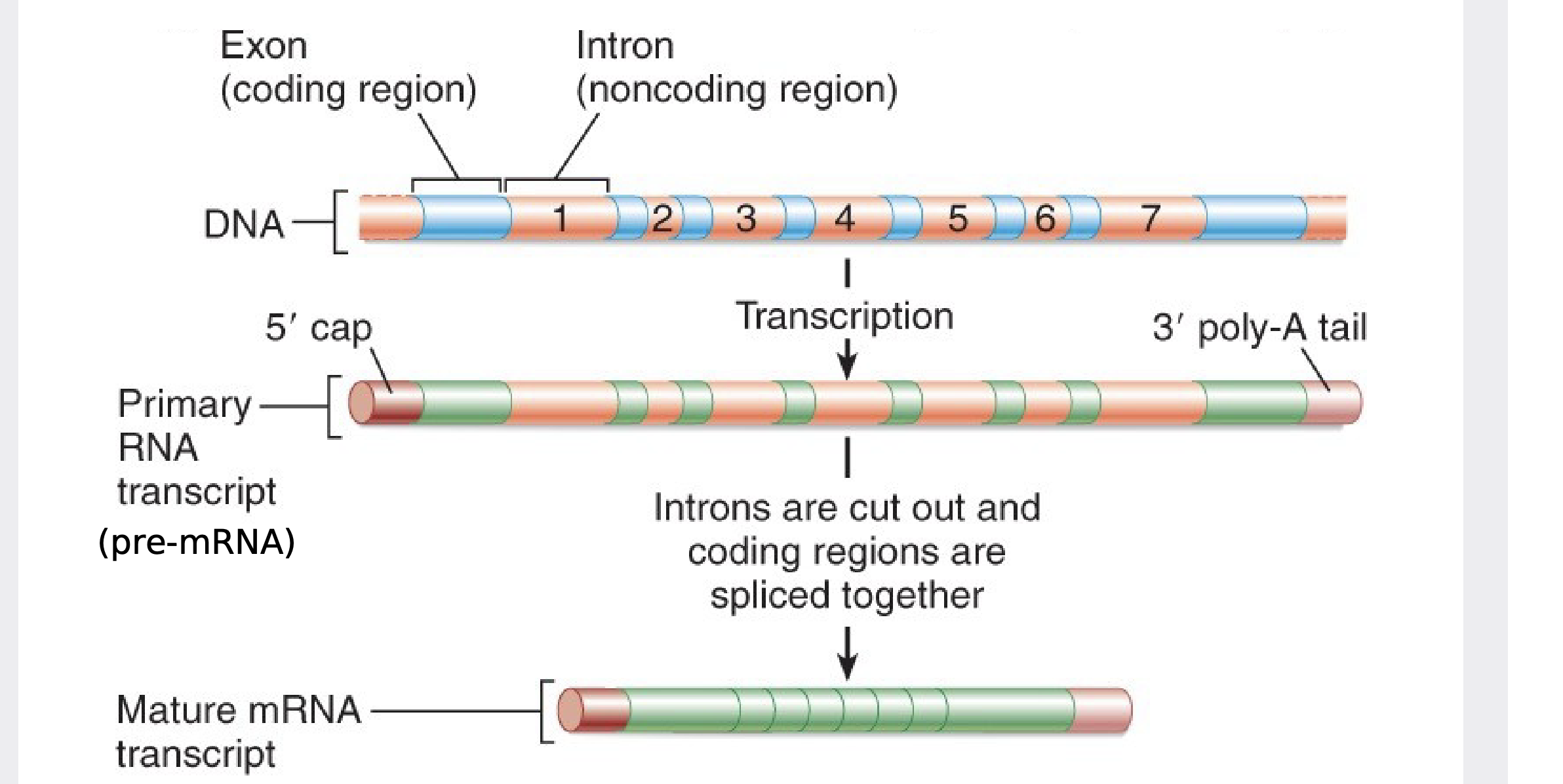
69
New cards
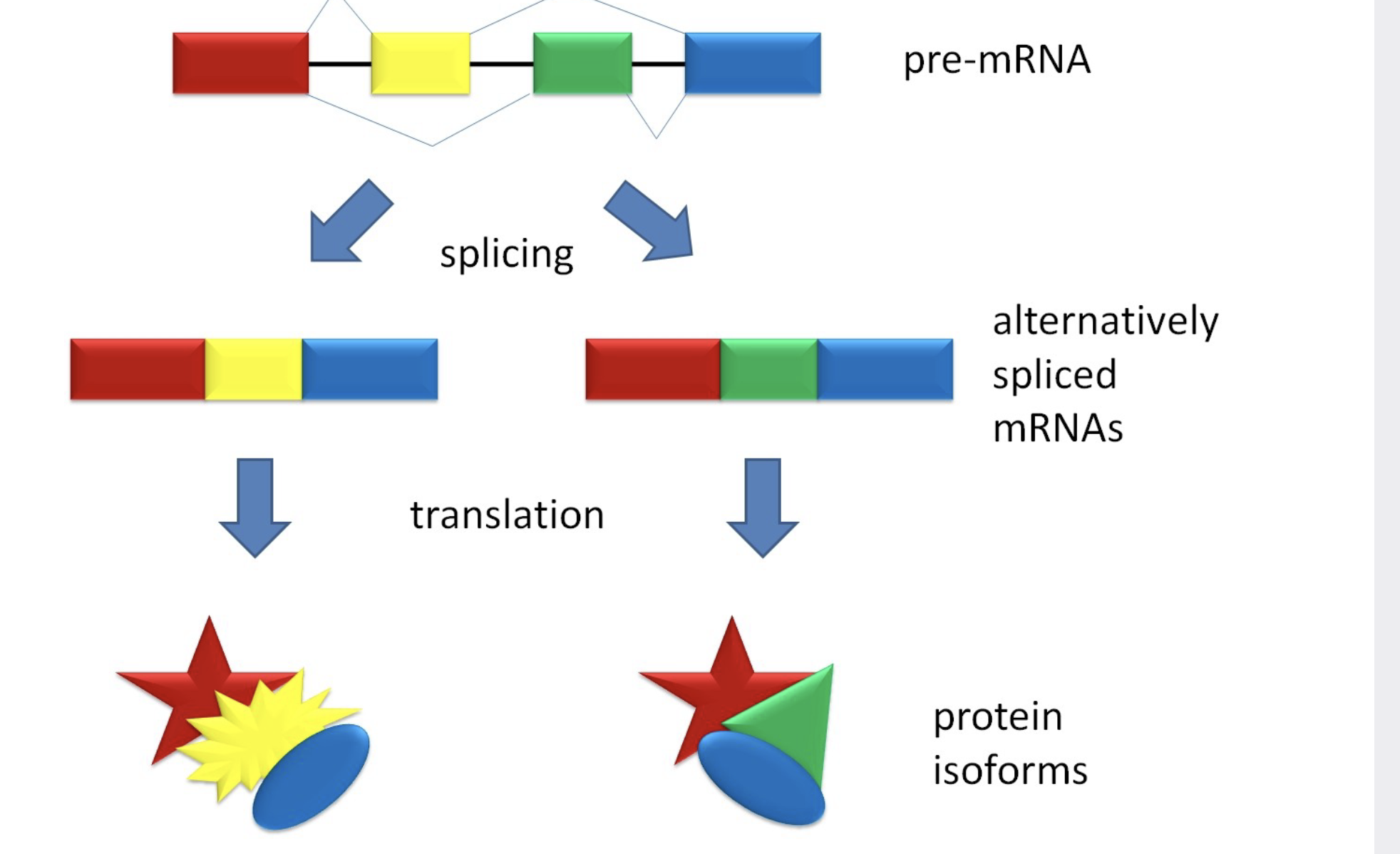
alternative splicing
Some introns contain sequences that regulate gene expression.
.Some genes can encode more than one kind of polypeptide, depending on which segments are \\n treated as exons during splicing. This is called alternative splicing.
.Consequently, the number of different proteins a eukaryotic organism can produce is much greater than its number of genes.
.Splicing can produce multiple forms of a protein from a single gene by joining together different combinations of exons.
.Some genes can encode more than one kind of polypeptide, depending on which segments are \\n treated as exons during splicing. This is called alternative splicing.
.Consequently, the number of different proteins a eukaryotic organism can produce is much greater than its number of genes.
.Splicing can produce multiple forms of a protein from a single gene by joining together different combinations of exons.
70
New cards
Eukaryotes mRNa to cytoplasm
.In a eukaryotic cell, the nuclear envelope separates transcription from translation.
.Eukaryotic RNA transcripts are modified through RNA processing to yield the finished mRNA which is exported from the nucleus.
.Eukaryotic RNA transcripts are modified through RNA processing to yield the finished mRNA which is exported from the nucleus.
71
New cards
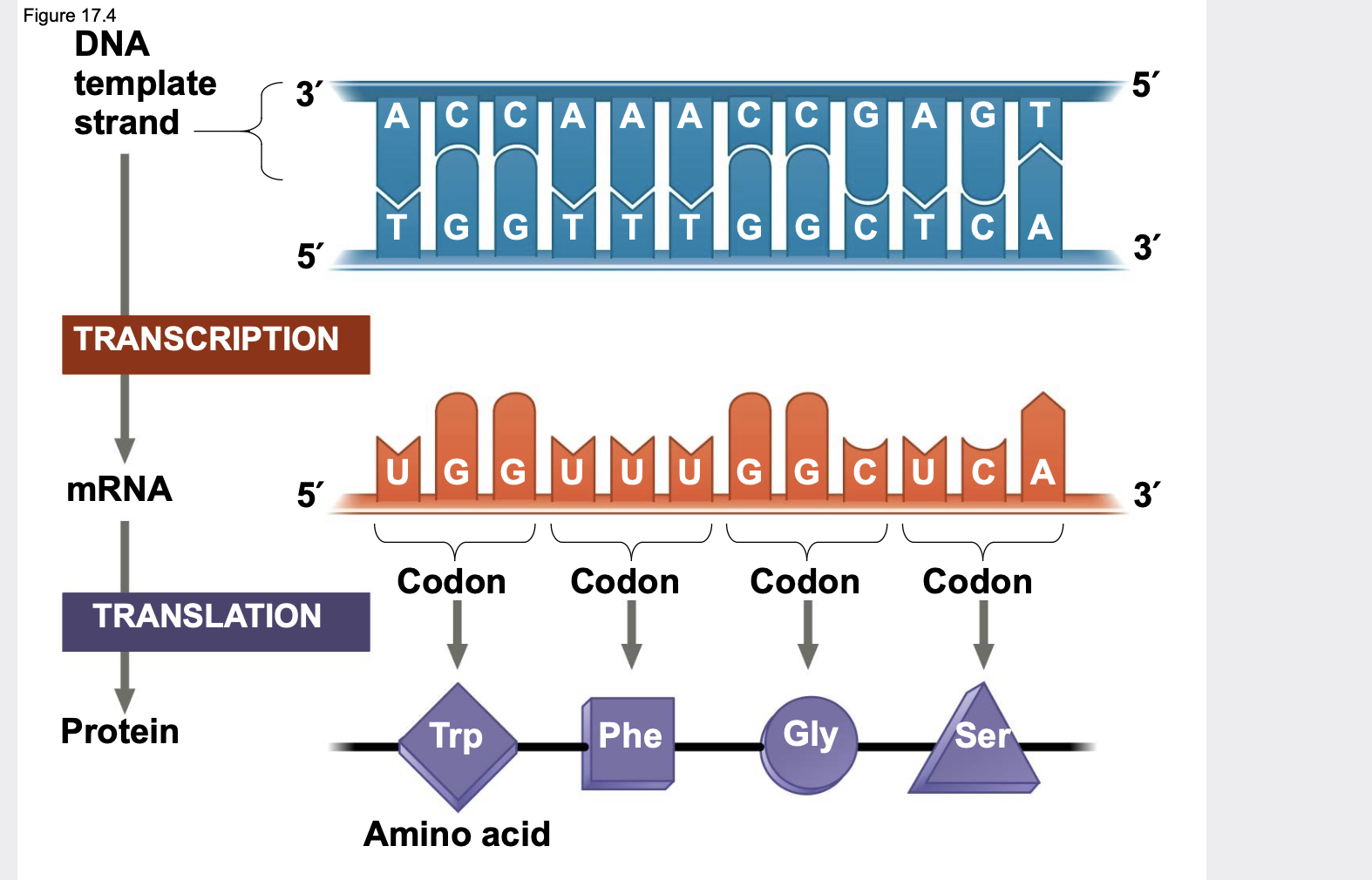
Translation: in codons
The flow of information from gene to protein is based on a triplet code.
.A codon is a triplet of bases, which codes for one amino acid.
.A codon is a triplet of bases, which codes for one amino acid.
72
New cards
The Genetic Code
the set of rules that decipher a triplet nucleotide sequence in RNA (codon) to an amino acid.
.Since there are 4 bases in DNA/RNA, a triplet can form 4 x 4 x 4= 64 possible combinations.
.Thus, there are 64 possible codons from 4 bases.
.Of the 64 codon triplets, – 61 code for amino acids (some amino acids are coded for by multiple codons) – 3 are stop codons, instructing the ribosomes to end translation.
.Since there are 4 bases in DNA/RNA, a triplet can form 4 x 4 x 4= 64 possible combinations.
.Thus, there are 64 possible codons from 4 bases.
.Of the 64 codon triplets, – 61 code for amino acids (some amino acids are coded for by multiple codons) – 3 are stop codons, instructing the ribosomes to end translation.
73
New cards
Translation
• Translation, the polymerization of amino acids into a protein, is catalyzed by ribosomes.
.A ribosome is a large complex of multiple proteins and several molecules of ribosomal RNA (rRNA).
.In addition to the ribosome, translation requires: – mRNA, which contains the genetic information copied from DNA and– transfer RNA (tRNA), another kind of RNAmolecule which decodes the genetic code
.A ribosome is a large complex of multiple proteins and several molecules of ribosomal RNA (rRNA).
.In addition to the ribosome, translation requires: – mRNA, which contains the genetic information copied from DNA and– transfer RNA (tRNA), another kind of RNAmolecule which decodes the genetic code
74
New cards
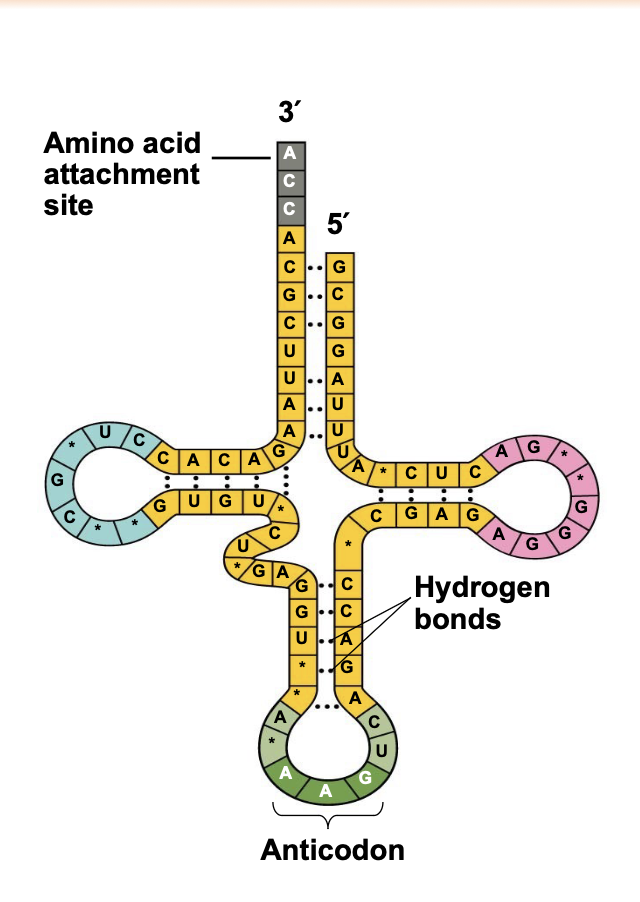
The Structure and Function of Transfer RNA
.tRNA decodes the genetic code \\n ▪ A tRNA contains a group of three bases called the anticodon which base-pair with a complementary codon on mRNA. \\n ▪ The 3 ́ end of a tRNA is covalently attached to the amino acid that its complementary codon calls for. A tRNA with an attached amino acid is called charged.
.Each codon in the mRNA thus has a corresponding tRNA with a complementary anticodon that brings the correct amino acid to the ribosome to be added to the growing polypeptide chain.
.A tRNA molecule consists of a single RNA strand that is only about 80 nucleotides long.
.The nucleotides within a tRNA form base-pairs with \n themselves to fold the tRNA into a specific 3D structure.
.Because of base-pairing throughout the molecule, tRNA twists and folds into a three-dimensional L- shaped structure
.Each codon in the mRNA thus has a corresponding tRNA with a complementary anticodon that brings the correct amino acid to the ribosome to be added to the growing polypeptide chain.
.A tRNA molecule consists of a single RNA strand that is only about 80 nucleotides long.
.The nucleotides within a tRNA form base-pairs with \n themselves to fold the tRNA into a specific 3D structure.
.Because of base-pairing throughout the molecule, tRNA twists and folds into a three-dimensional L- shaped structure
75
New cards
Ribosomes
Ribosomes facilitate pairing of tRNA anticodons with mRNA codons in protein synthesis \n ▪ Ribosomes are comprised of two subunits (small and large) which are both made of multiple proteins and ribosomal RNA (rRNA)
76
New cards
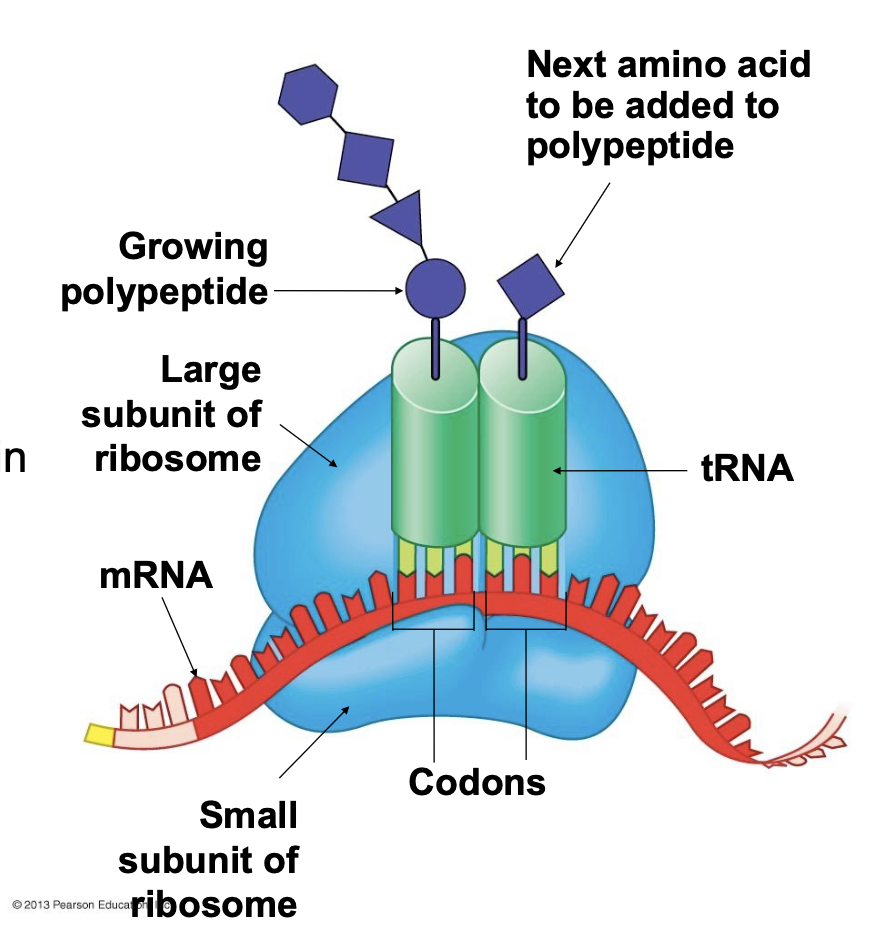
A ribosome during translation.
The ribosome positions the mRNA and tRNAs such that matching of anticodons in the tRNAs to the codons in the mRNA positions amino acids in the correct sequence to form a protein
77
New cards
Ribosome sites for tRNA
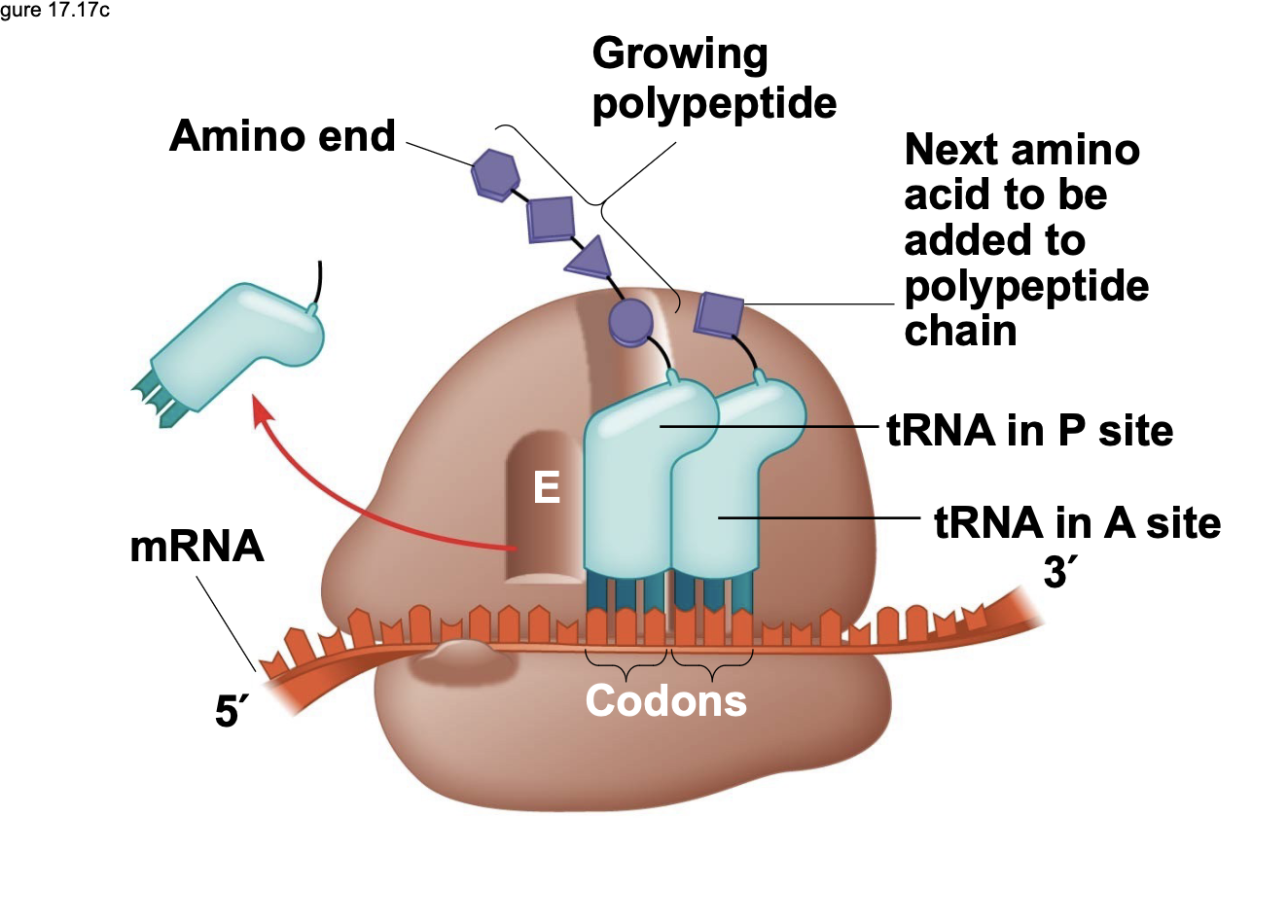
A ribosome has three binding sites for tRNA \n ▪ The P site holds the tRNA that has the growing polypeptide chain covalently attached to its 3 ́ end \n ▪ The A site holds the tRNA that has the next amino acid to be added to the chain attached to its 3 ́ end \n ▪ The E site is the exit site, where tRNAs that were previously in the P site will leave the ribosome after the growing polypeptide is transferred to the next tRNA
78
New cards
The three stages of translation
1\. Initiation
2\. Elongation
3\. Termination
.All three stages require protein “factors” that aid in the \\n translation process.
. Energy (ATP) is required for many steps also
2\. Elongation
3\. Termination
.All three stages require protein “factors” that aid in the \\n translation process.
. Energy (ATP) is required for many steps also
79
New cards
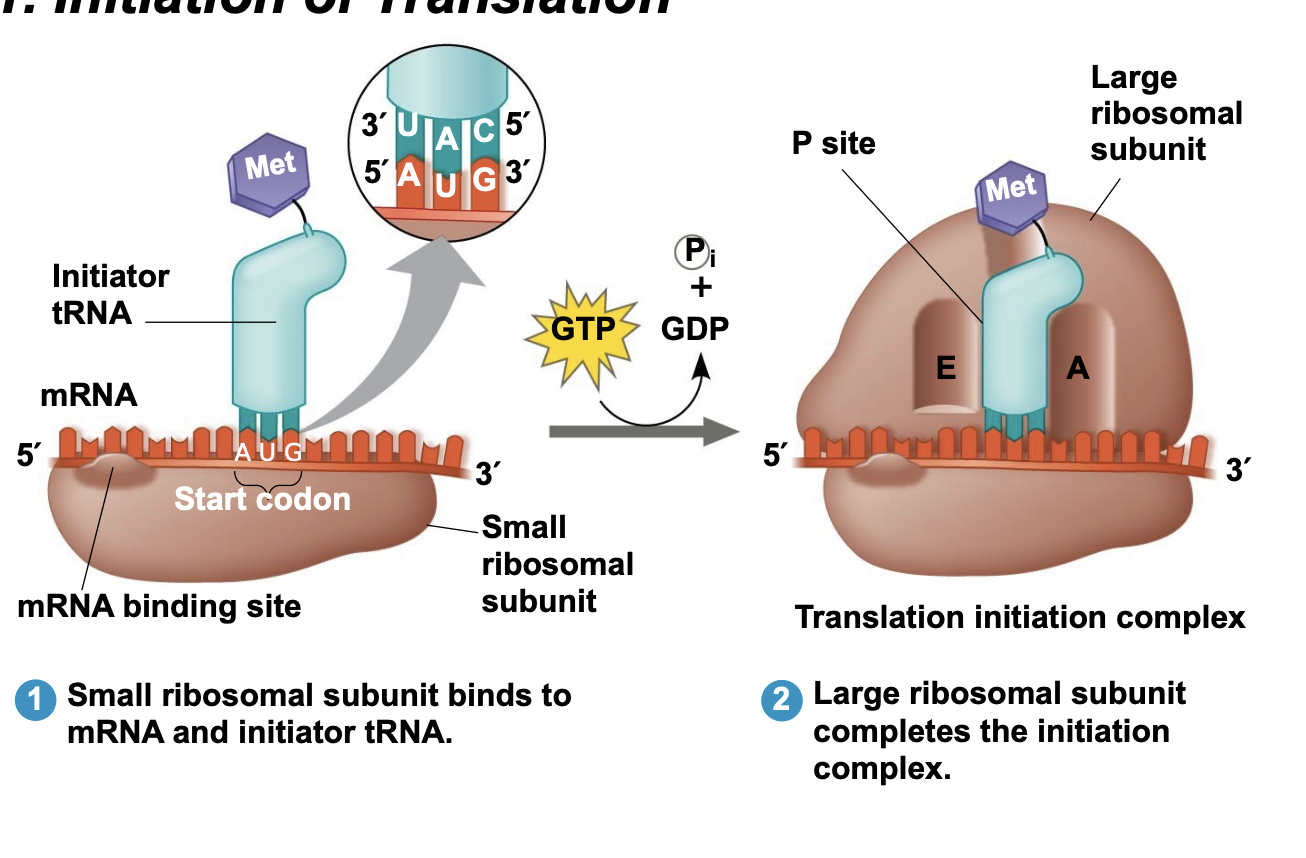
Initiation of Translation
.First, the small ribosomal subunit binds with mRNA and a special initiator tRNA.
.The anticodon of the initiator tRNA is complementary to and base pairs with the start codon (AUG), which codes for the amino acid methionine (Met).
.Proteins called initiation factors then bring in the large subunit to completes the translation initiation complex and place the first tRNA in the P site.
.The anticodon of the initiator tRNA is complementary to and base pairs with the start codon (AUG), which codes for the amino acid methionine (Met).
.Proteins called initiation factors then bring in the large subunit to completes the translation initiation complex and place the first tRNA in the P site.
80
New cards
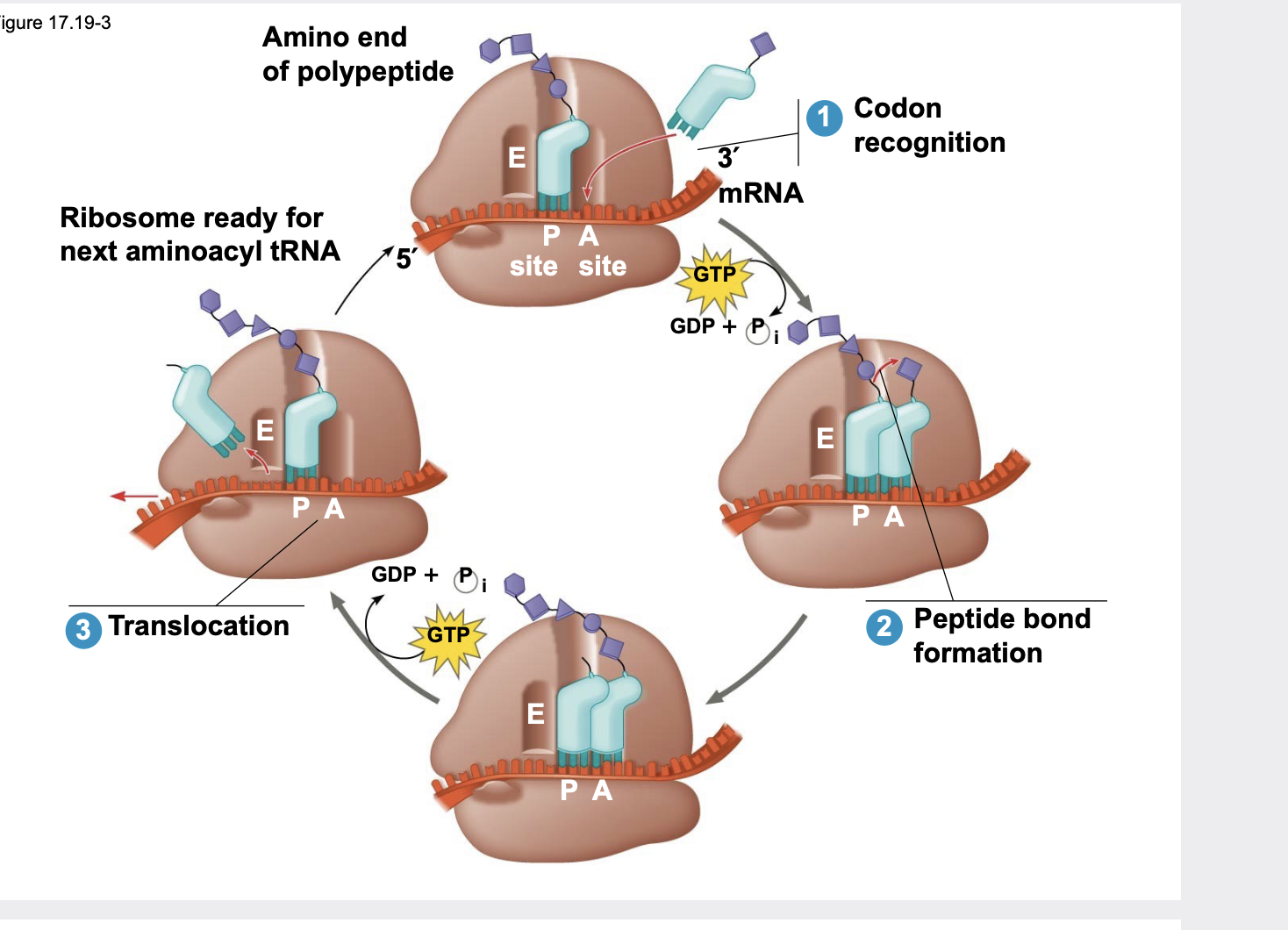
Elongation of translation
During elongation, amino acids are added one by one to the C-terminus of the growing polypeptide chain \\n .Each addition involves proteins called elongation factors and occurs in three steps:
1\. codon recognition,
2\. peptide bond formation
3\. translocation
.Translation proceeds along the mRNA in a 5′ → 3′ direction
1\. codon recognition,
2\. peptide bond formation
3\. translocation
.Translation proceeds along the mRNA in a 5′ → 3′ direction
81
New cards
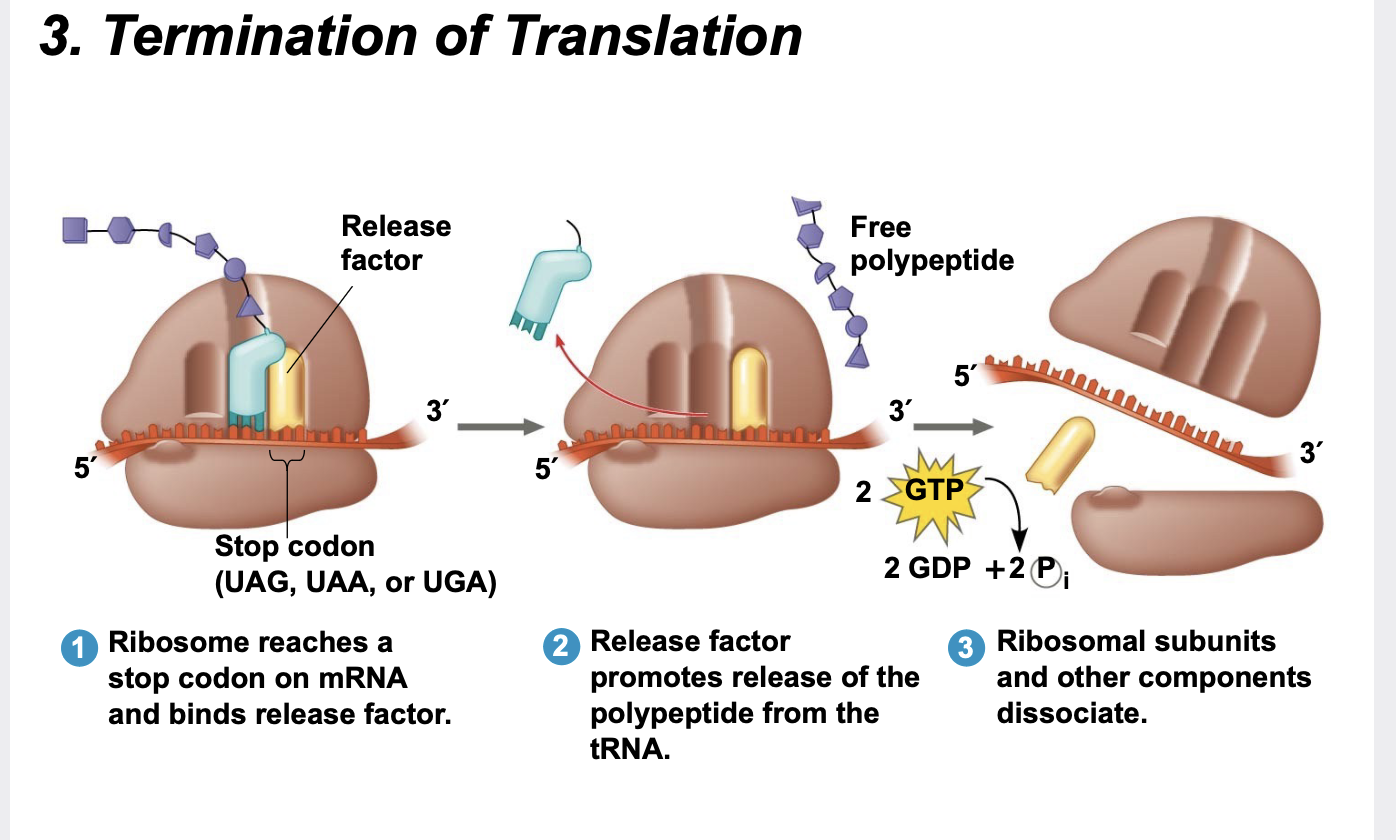
Termination of Translation
Termination occurs when a stop codon (UAA, UAG, or UGA) in the mRNA reaches the A site of the ribosome.
The A site accepts a protein called a release factor.
The release factor causes the addition of a water molecule to the C-terminus of the growing polypeptide which releases it from the tRNA.
. After polypeptide release, the entire translation assembly (ribosome, tRNAs, mRNA, etc.) comes apart.
.The mRNA can be translated again and again.
The A site accepts a protein called a release factor.
The release factor causes the addition of a water molecule to the C-terminus of the growing polypeptide which releases it from the tRNA.
. After polypeptide release, the entire translation assembly (ribosome, tRNAs, mRNA, etc.) comes apart.
.The mRNA can be translated again and again.
82
New cards
Populations and locations of ribosomes
Two populations of ribosomes are evident in cells: free ribosomes (in the cytosol) and bound ribosomes (attached to the ER).
Free ribosomes mostly synthesize proteins that function in the cytosol \n ▪ Bound ribosomes make proteins of the endomembrane system and proteins that are secreted from the cell.
.Ribosomes can switch back and forth from free to bound, depending on which mRNA they are translating
Free ribosomes mostly synthesize proteins that function in the cytosol \n ▪ Bound ribosomes make proteins of the endomembrane system and proteins that are secreted from the cell.
.Ribosomes can switch back and forth from free to bound, depending on which mRNA they are translating
83
New cards
Making Multiple Polypeptides
.Multiple ribosomes can translate a single mRNA simultaneously, forming a polyribosome (or polysome).
Polyribosomes enable a cell to make many copies of a polypeptide very quickly
Polyribosomes enable a cell to make many copies of a polypeptide very quickly
84
New cards
Proteins don’t last forever.
.Proteins are eventually broken down by the cell. This is an important part of controlling their activity.
.Proteins are hydrolyzed by a complex called the proteasome which breaks down the polypeptides into smaller peptides and eventually amino acids.
.Proteins are marked for destruction by the addition of a protein called ubiquitin (Ub). Multiple ubiquitin proteins are added to the target protein in a chain. This is called polyubiquitination.
.Polyubiquitinated proteins are recognized by the proteasome and hydrolyzed.
.Proteins are hydrolyzed by a complex called the proteasome which breaks down the polypeptides into smaller peptides and eventually amino acids.
.Proteins are marked for destruction by the addition of a protein called ubiquitin (Ub). Multiple ubiquitin proteins are added to the target protein in a chain. This is called polyubiquitination.
.Polyubiquitinated proteins are recognized by the proteasome and hydrolyzed.
85
New cards
Mutations of one or a few nucleotides can affect protein structure and function
Mutations are changes in the DNA.
Point mutations are changes in just one base pair of a gene.
.A point mutation can, but does not always, lead to the production of an abnormal protein.
Point mutations are changes in just one base pair of a gene.
.A point mutation can, but does not always, lead to the production of an abnormal protein.
86
New cards
Types of point mutations
Point mutations are changes in a single nucleotide. \n There are three kinds of point mutations: \n 1. Silent mutations have no effect on the amino acid sequence because they change one codon into another codon that codes for the same amino acid. There is no effect on the protein since the amino acid sequence is not changed. \n 2. Missense mutations change the amino acid sequence because they change one codon into another codon that codes for a different amino acid. Sometimes they have little to no effect on a protein’s function because the changed amino acid is very similar to the original amino acid. This is called a conservative change. \n 3. Nonsense mutations shorten the amino acid sequence because they change a codon into a stop codon. This nearly always leads to a too short, nonfunctional protein.
87
New cards
Insertions and Deletions
▪ Insertions and deletions are additions or losses of one or more nucleotides in a gene.
These mutations are more likely to have a disastrous effect on the resulting protein than point mutations do.
.Insertion or deletion of nucleotides may alter the reading frame, producing a frameshift mutation.
.In a frameshift mutation, the nucleotides in the mRNA are grouped into codons differently, causing extensive changes in the amino acid sequence and usually a \\n nonfunctional protein.
These mutations are more likely to have a disastrous effect on the resulting protein than point mutations do.
.Insertion or deletion of nucleotides may alter the reading frame, producing a frameshift mutation.
.In a frameshift mutation, the nucleotides in the mRNA are grouped into codons differently, causing extensive changes in the amino acid sequence and usually a \\n nonfunctional protein.
88
New cards
Mutations in noncoding DNA
Mutations in DNA sequences that control gene expression could lead to misregulation, and in turn, loss of fitness
89
New cards
Differential Expression of Genes
.Prokaryotes and eukaryotes precisely regulate gene expression to maintain homeostasis and to respond to environmental conditions.
.In multicellular eukaryotes, gene expression regulates development and is responsible for differences in cell types.
.RNA molecules play many roles in regulating gene expression in eukaryotes
.In multicellular eukaryotes, gene expression regulates development and is responsible for differences in cell types.
.RNA molecules play many roles in regulating gene expression in eukaryotes
90
New cards
Bacteria often respond to environmental change by regulating transcription
Bacteria control gene expression primarily by controlling how much each of their genes are transcribed.
.One mechanism for control of gene expression in bacteria is the operon model.
.An operon is a group of genes that are always expressed together.
.One mechanism for control of gene expression in bacteria is the operon model.
.An operon is a group of genes that are always expressed together.
91
New cards
Operons
.A group of functionally related genes can be simultaneously controlled by a single “switch”.
.The “switch” is a sequence of DNA called an operator, which usually positioned within the promoter of the first gene in the group
.An operon is the entire stretch of DNA that includes the operator, the promoter, and the group of genes that they simultaneously control.
.Genes whose expression is regulated together are said to be co-expressed.
.The trp operon allows the expression of all the genes involved in synthesizing the amino acid tryptophan to be expressed at the same time.
.The “switch” is a sequence of DNA called an operator, which usually positioned within the promoter of the first gene in the group
.An operon is the entire stretch of DNA that includes the operator, the promoter, and the group of genes that they simultaneously control.
.Genes whose expression is regulated together are said to be co-expressed.
.The trp operon allows the expression of all the genes involved in synthesizing the amino acid tryptophan to be expressed at the same time.
92
New cards
Controlling expression of an operon
.An operon can be switched off by a repressor, a protein that binds to the operator
.The repressor prevents transcription by binding to the operator and blocking RNA polymerase from binding to the promoter.
.A separate gene, outside the operon, encodes the repressor protein
.A gene (or operon) can be controlled by a combination of both positive regulators (activators) and negative regulators (repressors).
.The repressor prevents transcription by binding to the operator and blocking RNA polymerase from binding to the promoter.
.A separate gene, outside the operon, encodes the repressor protein
.A gene (or operon) can be controlled by a combination of both positive regulators (activators) and negative regulators (repressors).
93
New cards
Repressible and Inducible Operons: Two Types of Negative Gene Regulation
.Repressors are negative regulators. When activated, repressors bind DNA and block RNA polymerase, preventing transcription.
.A repressible operon is one that is transcribed unless a molecule activates a repressor which then turns it off.
• The repressor is usually inactive and allows transcription. Once activated, the repressor binds to the operator and shuts off (represses) transcription.
.An inducible operon is one that is not transcribed unless a molecule called an inducer inactivates the repressor and thus turns on transcription.
• The repressor is usually active and blocks transcription. The inducer inactivates the repressor, turning on (inducing) transcription.
.A repressible operon is one that is transcribed unless a molecule activates a repressor which then turns it off.
• The repressor is usually inactive and allows transcription. Once activated, the repressor binds to the operator and shuts off (represses) transcription.
.An inducible operon is one that is not transcribed unless a molecule called an inducer inactivates the repressor and thus turns on transcription.
• The repressor is usually active and blocks transcription. The inducer inactivates the repressor, turning on (inducing) transcription.
94
New cards
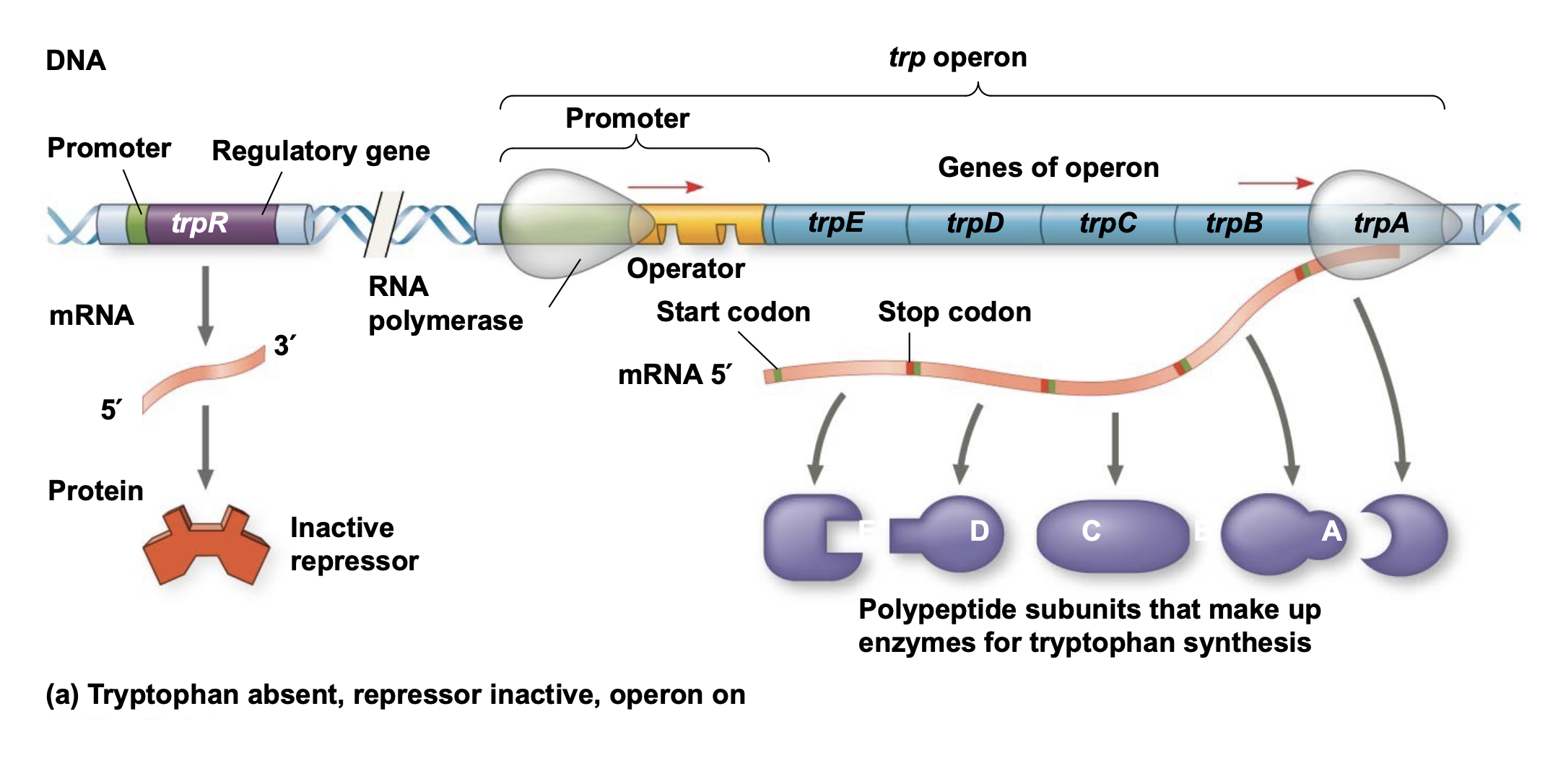
The trp operon is repressible.
.The trp repressor protein requires tryptophan to bind DNA \n and thus turn off transcription of the trp operon.
.When there is no (or very low) tryptophan, the repressor is \n inactive (cannot bind DNA), allowing RNA polymerase to bind \n the promoter and transcribe the genes in the trp operon.
.When tryptophan is present, it binds to and activates the trp \n repressor protein which then binds the operator and turns off \n transcription.
.Thus the trp operon is turned off (repressed) if tryptophan levels are high and turned on when tryptophan levels are low.
.When there is no (or very low) tryptophan, the repressor is \n inactive (cannot bind DNA), allowing RNA polymerase to bind \n the promoter and transcribe the genes in the trp operon.
.When tryptophan is present, it binds to and activates the trp \n repressor protein which then binds the operator and turns off \n transcription.
.Thus the trp operon is turned off (repressed) if tryptophan levels are high and turned on when tryptophan levels are low.
95
New cards
The lac operon
an inducible operon and contains genes that code for enzymes used in the metabolism \\n of lactose.
.By default, the lac repressor is active and switches the lac operon off.
.A molecule called an inducer inactivates the repressor to turn the lac operon on.
. Inducers of the lac operon include lactose and other molecules very similar in structure to lactose.
.By default, the lac repressor is active and switches the lac operon off.
.A molecule called an inducer inactivates the repressor to turn the lac operon on.
. Inducers of the lac operon include lactose and other molecules very similar in structure to lactose.
96
New cards
Activator: positive gene regulation
Another type of gene regulator protein, called an activator, can increase transcription.
.when an activator is activated, it binds DNA and helps RNA polymerase bind to a promotor, increasing transcription.
.when an activator is activated, it binds DNA and helps RNA polymerase bind to a promotor, increasing transcription.
97
New cards
Eukaryotic gene expression
.all organisms must regulate which genes are expressed at any given time.
.In multicellular organisms regulation of gene expression is essential for cell specialization and development
.In multicellular organisms regulation of gene expression is essential for cell specialization and development
98
New cards
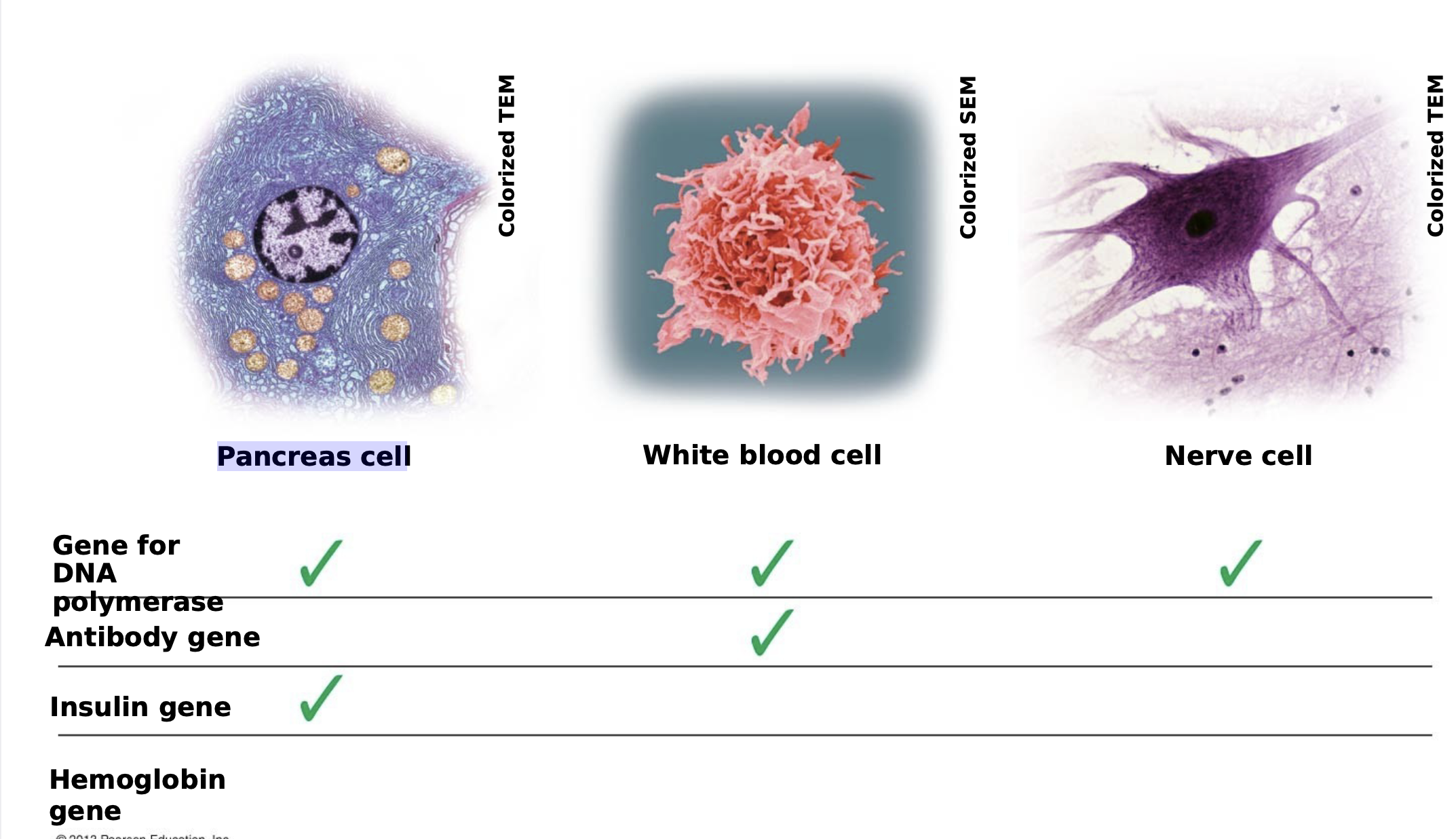
Differential Gene Expression
.Almost all the cells in a multicellular organism are genetically identical.
.Differences between cell types result from differential gene expression, the expression of different genes by cells with the same genome.
.Abnormalities in gene expression can lead to diseases including cancer.
.Gene expression is regulated at many stages
.Differences between cell types result from differential gene expression, the expression of different genes by cells with the same genome.
.Abnormalities in gene expression can lead to diseases including cancer.
.Gene expression is regulated at many stages
99
New cards
Differences in gene expression lead to different cell types
Pancreas cell: Gene for DNA, insulin gene
White blood cell: Gene for DNA, polymerase antibody gene
Nerve Cell: Gene for DNA
White blood cell: Gene for DNA, polymerase antibody gene
Nerve Cell: Gene for DNA
100
New cards
embryonic development and gene expression
.During embryonic development, a fertilized egg gives rise to many different cell types.
Cell types are organized successively into tissues, organs, organ systems, and the whole organism.
.Gene expression orchestrates the developmental programs of animals.
Cell types are organized successively into tissues, organs, organ systems, and the whole organism.
.Gene expression orchestrates the developmental programs of animals.