L11 Genes, Genomes, and transposable elements
1/43
Earn XP
Description and Tags
L11
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
44 Terms
genome
the entirety of an organism’s hereditary info
usually DNA
what is the eukaryotic genome composed of?
coding DNA (islands)
and
non-coding DNA (open ocean)
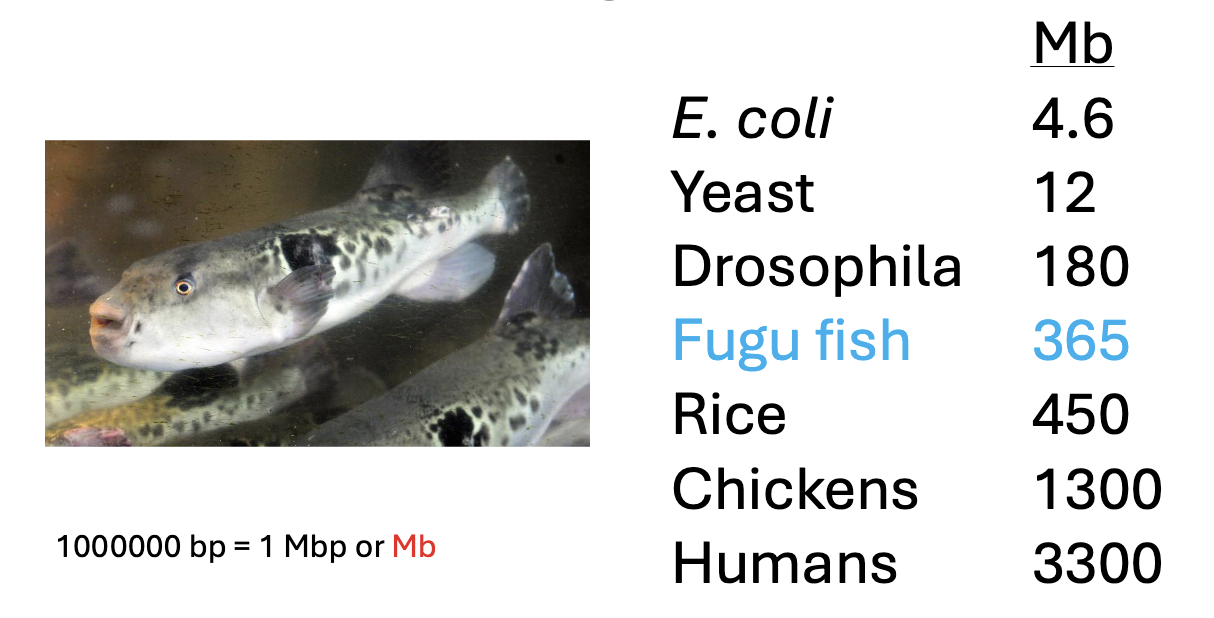
does genome size correlate to genome complexity? what account for diffce in genome size?
not necessarily… the difference in genome size is largely due to the amount of non-coding DNA
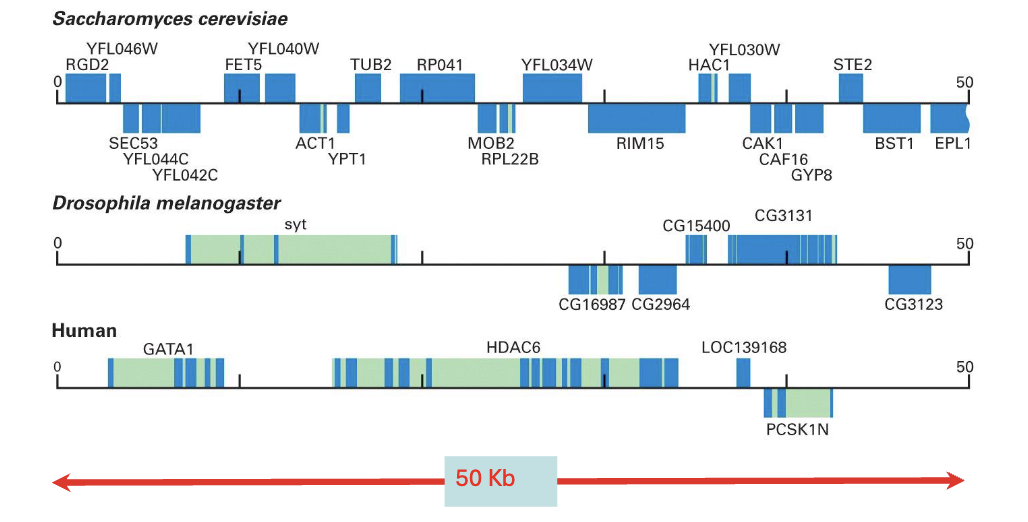
what leads to gene density variability among eukaryotes?
the size of intragenic (i.e. introns) and intergenic regions (i.e. between genes)
how much of the human genome do coding sequences account for ?
abt 3%
gene
the entire nucleic acid sequence that is necessary for the synthesis of a functional product (polypeptide or RNA)
genes are transcribed
what do exons of a gene contain
the coding region or Open Reading Frame (ORF)
contains codons telling ribosome to start/stop translation — aka protein synthesis
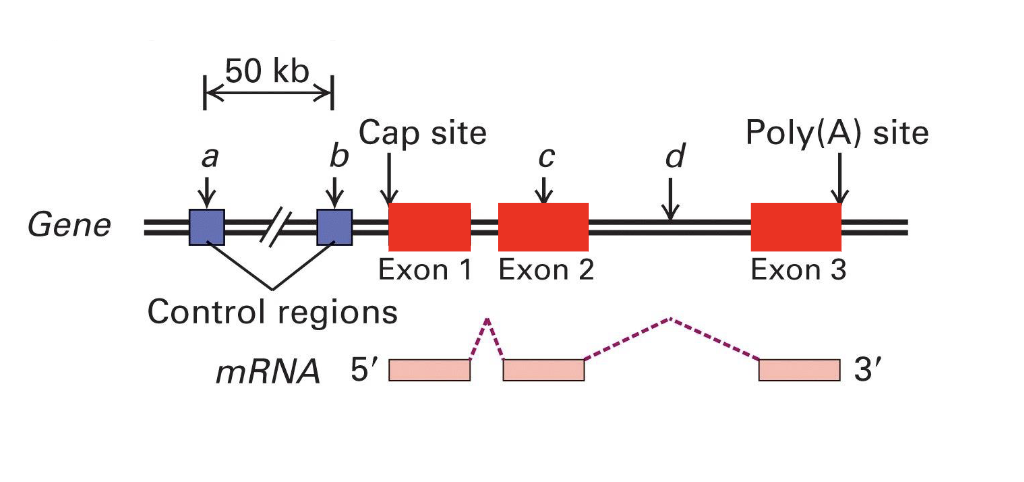
control regions
contain promoter and other cis- regulatory factors (like enhancers and silencers)
are found either upstream or downstream to coding regions
genes need these to control regions for transcription to occur
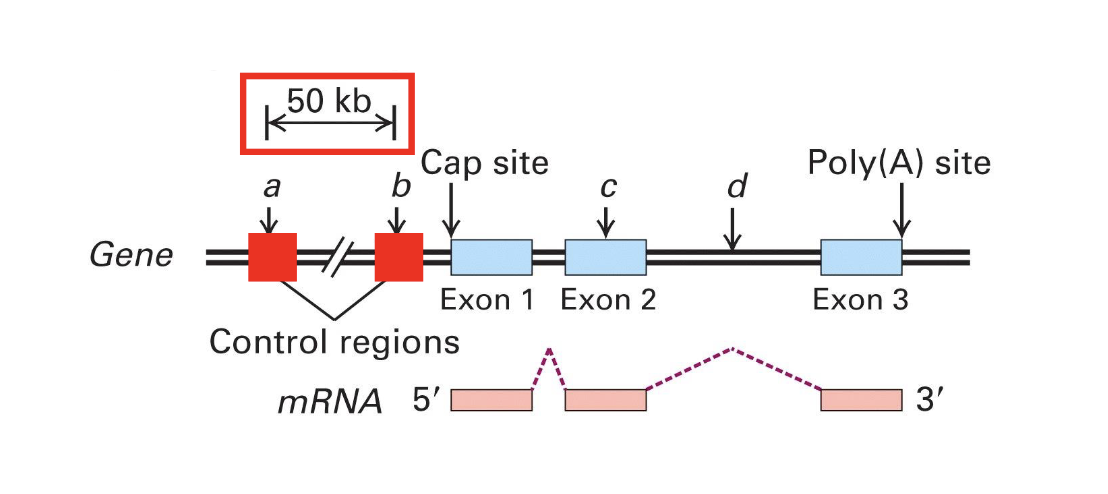
introns
separe the exons and are spliced out during mRNA processing
aren’t used for the final product
transcription unit
a region in DNA bounded by an initiation (start) site and termination site that is transcribed into a single primary transcript
aka the sequence that is actually copied in RNA
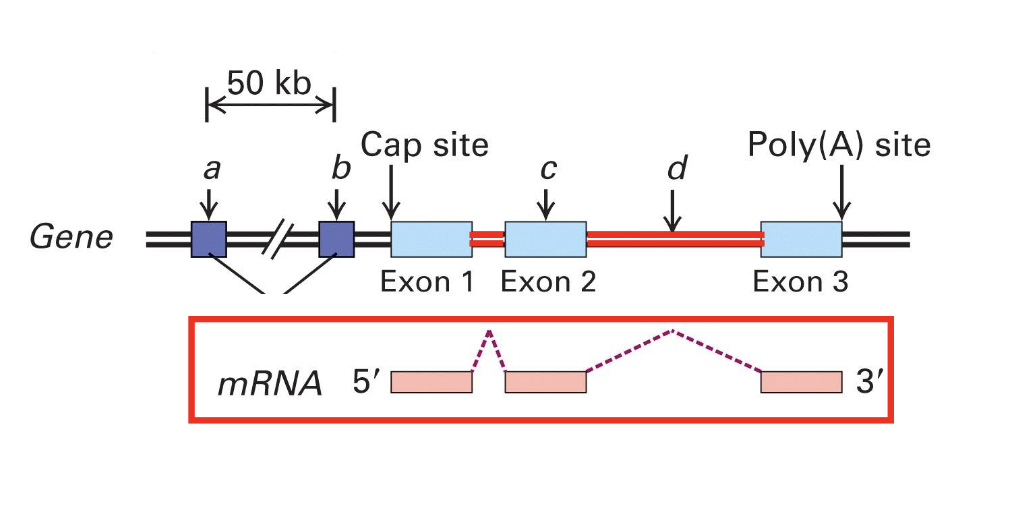
introns can vary in size. what can we say about them in human genes?
many human genes >10kB in length
some are over 1Mbp
vast majority (>95%) of an average human gene is non-coding
mostly intron sequences
represent about 42% of the total length in the genome total
what is the function on introns
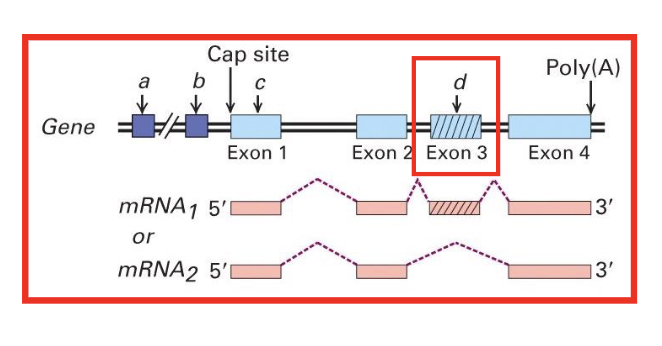
eukaryotic genes are alternatively transcribed and processed
generates multiple difft transcripts from the same gene
isoforms
the multiple forms of a protein produced by alternative splicing
solitary or single copy genes
25-50% of protein-coding genes
represented once in the genome
gene family
made up from a set of related genes formed by duplication of an original single-copy gene
called duplicates, occur in multiple copies
what can we say about the sequences of protiens with similar functions
they often contain similar AA sequences that encode functional domains
unlikely that this would have been generated independently => they must come from the same gene
what is an example of a sequence alligment technique we can use to find nucleic acid and protein sequence similarity
BLAST
what is the difference among genome size in species due to ?
mostly due to difft amts of non-coding DNA and Transposable elements
complexity cannot be explained by number of genes
gene duplication
an imp process in evolution
new gene copies either
evolve a new function
degenerate over time and lose their function => become pseudogenes
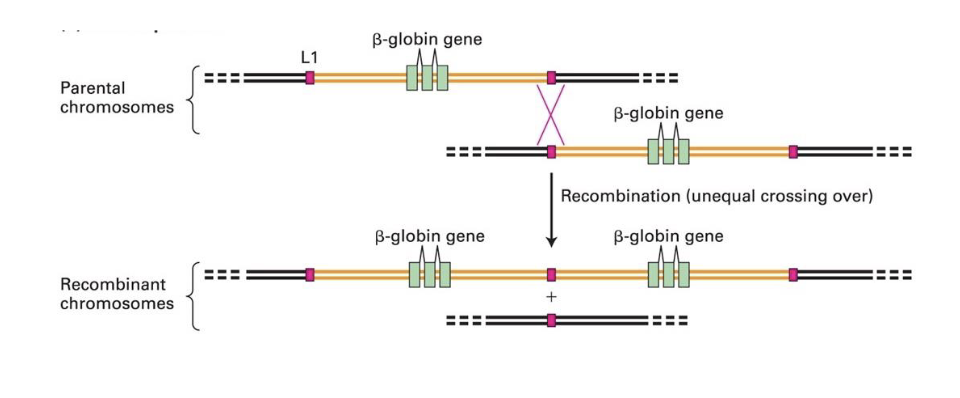
what does the comparison of related protein seqs in difft spp illuminate?
the evo relationships between these
orthologs
the same protein in different species (α-tubulin in humans and flies)
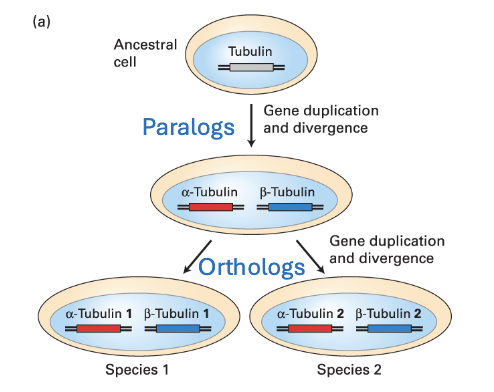
Paralogs
closely related proteins in the same species (α-tubulin and β-tubulin
in humans)
intragenic non-coding DNA
includes
introns
are spliced out
UTRS (untranslated regions)
are part of mature mRNA but not translated
Simple Sequence Repeats SSrs
6% of the genome
first group part of non-coding DNA
includes
minisatellite DNA
microsatellite DNA
minisatelite DNA (an SSR)
Repeat units are ≈14 to 100 bp in length
20-50 tandem repeat units
Arrays of 1 to 5 kbp in length
Often in centromeres and telomeres
microsatellite DNA (an SSR)
Repeat units are typically 1 to 4 bp in length
smaller than minisatellite DNA
Arrays of up to ≈600 bp in length and composed of tandem repeat units
Sometimes found in transcription units
Expansion underlie several neuromuscular diseases
like myotonic dystrophy
and spinocerebellar ataxia
how does slippage occur in DNA?
during the replication of long repeats
polymerase stalls
slips or looses its place on the template strand
causes the template and synthesized strand to missallign leading to temporary loop formation
synthesis resumes
the restabilization can result in the addition of more repeats than were in the OG template — this is called repeat extension
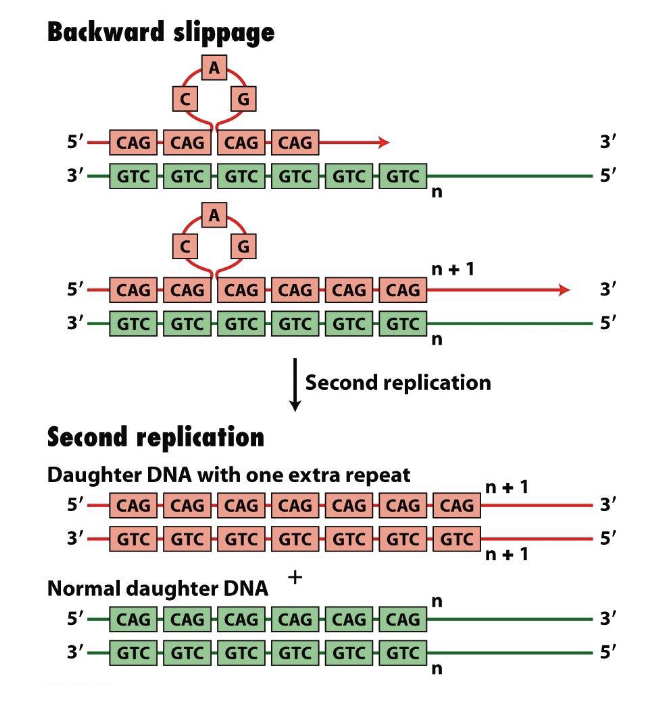
how is gene slippage associated to neuromuscular diseases?
e.g. Huntington’s
results in the productuion of protein that form toxic aggregates in neuronal cells
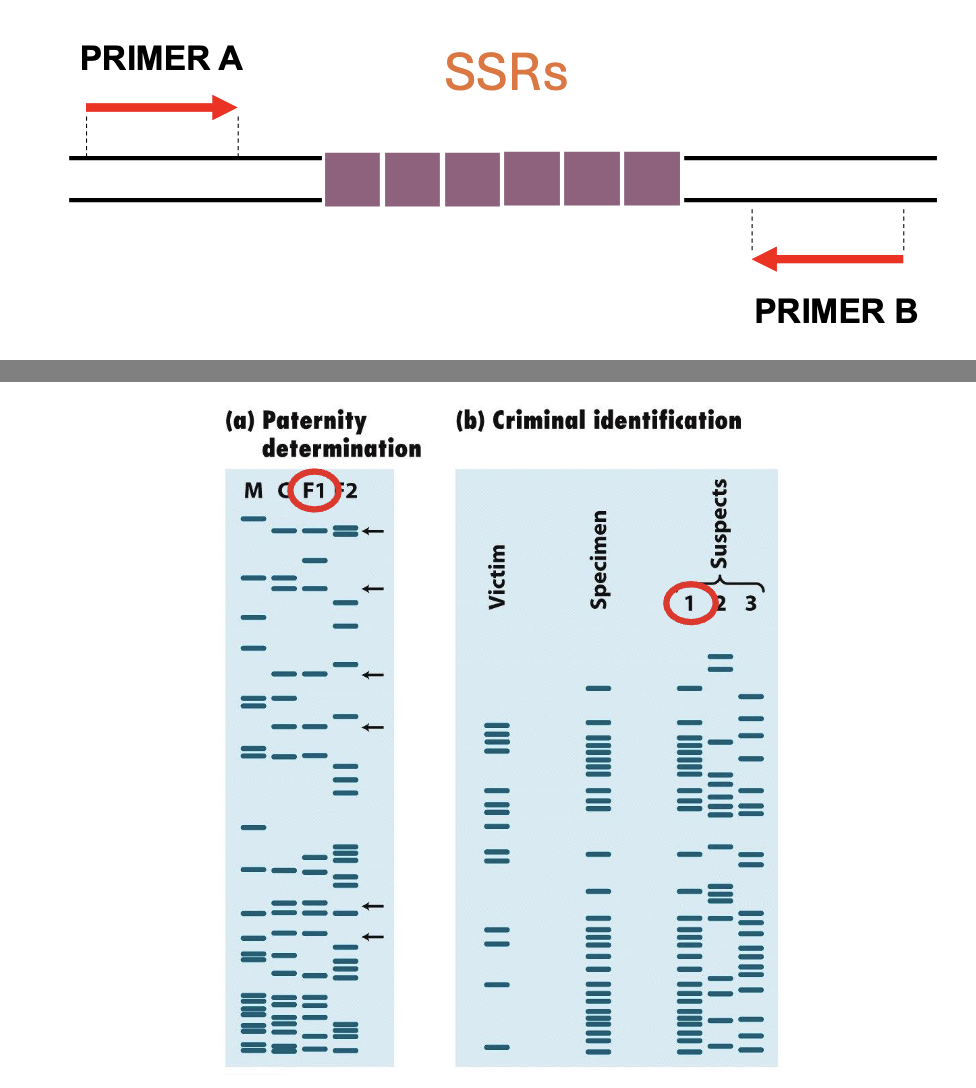
how can the hypervariable nature of SSRs be exploited for DNA fingerprinting protocols? (for paternity determination or criminal identification)
SSRs can be amplified by PCR ( or studied by southern blot)
the n° of repeats is determined by high res gel electrophoresis
what are the 2 major classes of Transposable Elements (TEs) we studied?
DNA Transposons
3% of genome
just jump from one position to another
Retrotransposons
40% of genome
copy themselves threough an RNA intermediate before moving around (that’s why there are more)
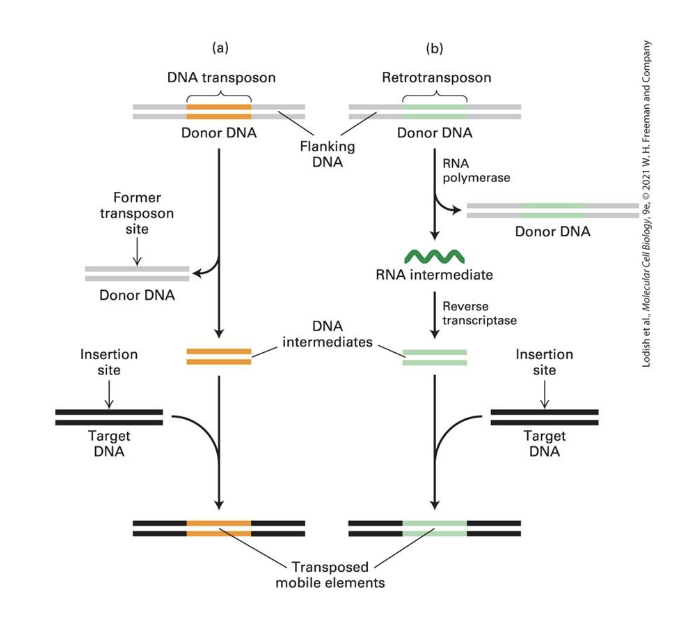
Transposable (Mobile) DNA Elements aka Jumping Genes (what do they do? what is their impact? who identified them?)
move within the genomes by difft mechanisms
influenced evolution
can cause mutations leading to disease
originally identified by Barabara McClintock by studying coor pattern formation in maize (cause by production of the pigment anthocyanin)
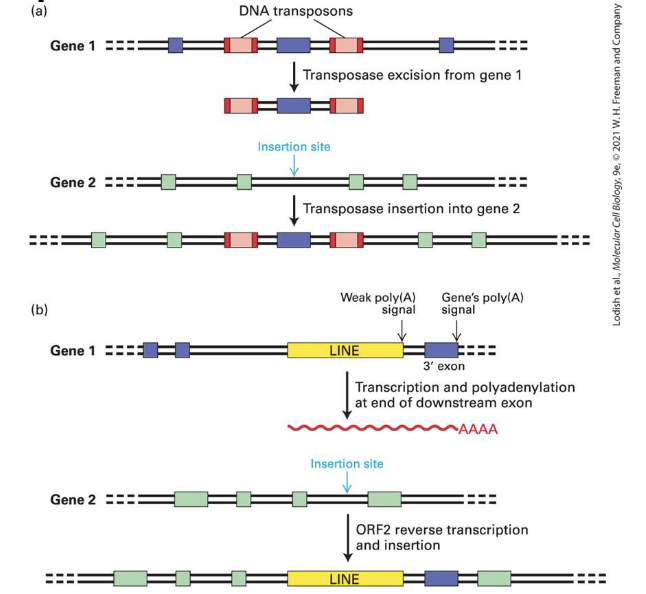
what is the mechanism of transposition
DNA transposases inserted through cut-and-paste mechanism
(1)
Transposase makes blunt ended cuts in donor DNA
and staggered cuts in taget DNA
(2)
transposase ligates IS10 to 5’ single stranded ends of target DNA
(3)
cellular DNA polymerase extends 3’ cut ends and ligase joins extended 3’ ends to IS10 5’ ends
ligating the blunt-end transposon sequence in the staggered-ended recipient seaquence reslts in a short duplication of DNA (9bp)
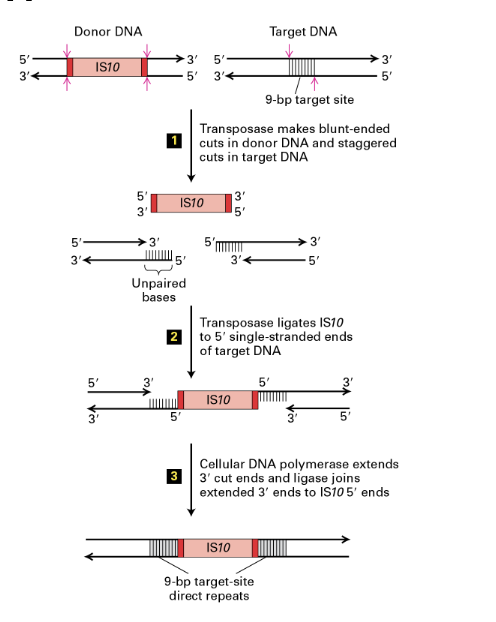
Transposase
enzyme that catalyzes the insertion of DNA transposons
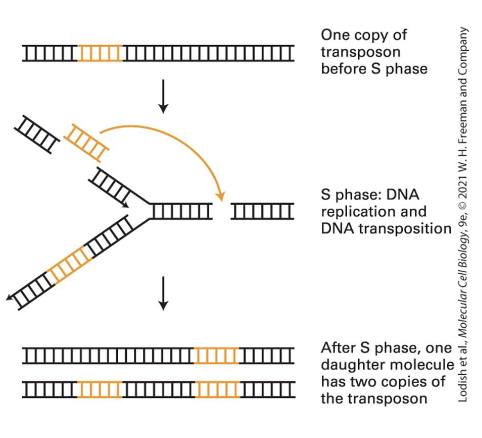
how can a DNA transposon increase its copy number (despite its cut-and-paste mechanism)
during S phase
if transposon moves from region that has replicated to one that has not then the copy number will increase by one in one of the daughter chromosomes (one daughter molecule will have two copies fo the tranposon)
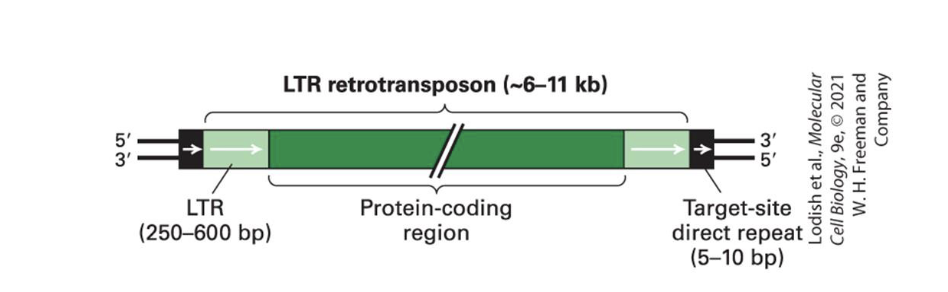
LTR (Long Terminal Repeats) - a type of retrotransposon
inserted at 440,000 sites, abt 8% of our genomes
are similar to retroviruses but lack envelope proteins
their protein coding region encodes
reverse transcriptase
integrase (similar function to transposase of DNA transposons)
and other proteins
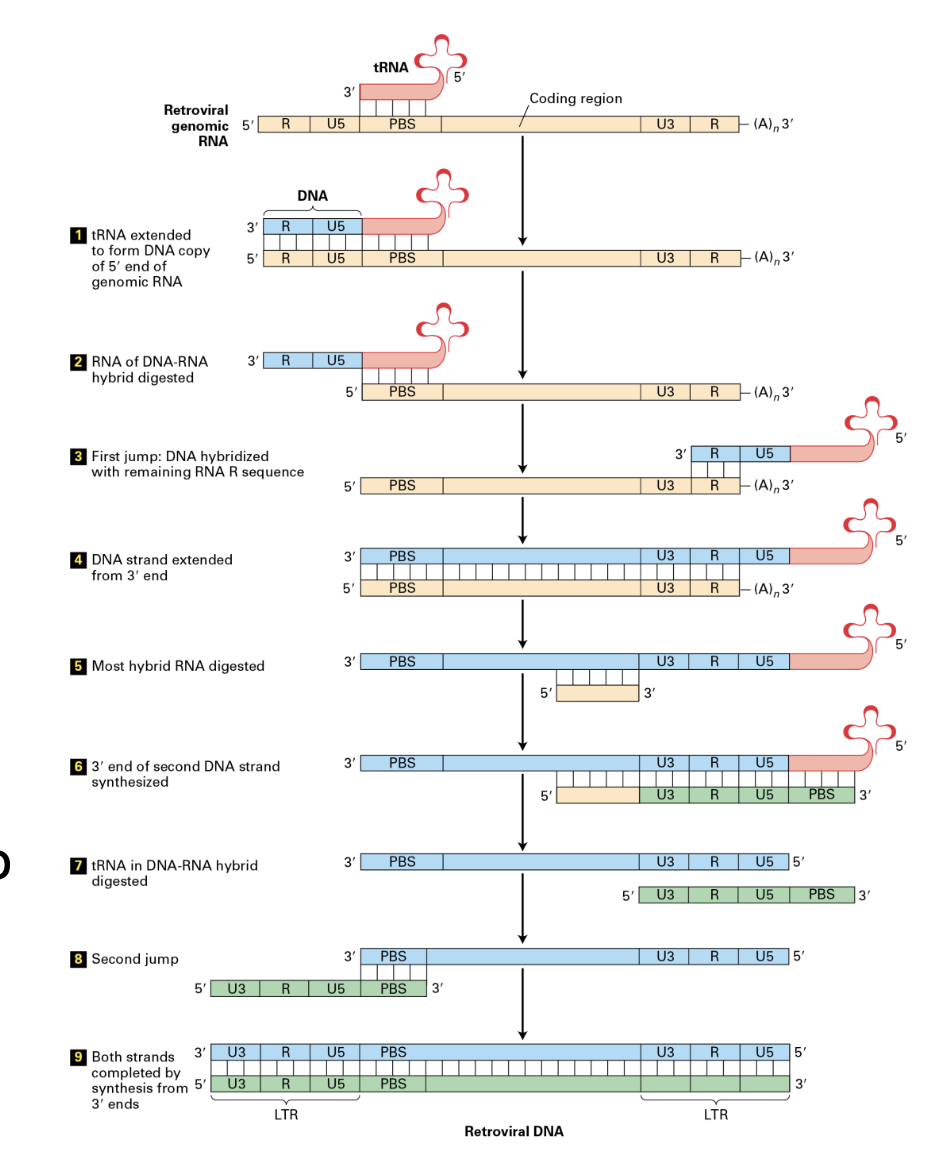
mechanism of copy-and-paste of LTRs (3 steps)
LTRs are first transcribed
generating an RNA copy of most of their sequence, excluding part of the ends
LTRs require a retrotranscriptase to convert the RNA molecule into DNA through a multiple step process.
This process occur in the cytoplasm.
A molecule of tRNA is used as a primer in the process
RNA is complementary to tRNA which is why it can be used as a primer
DNA is then imported to the nucleus in complex with integrase (a protein related to transposases used by DNA transposons)
Ingrase will mediate the insertion into the genome using a similar mechanism as transposases do for DNA transposons
what are two types of nonviral retrotransposons
LINEs
SINEs
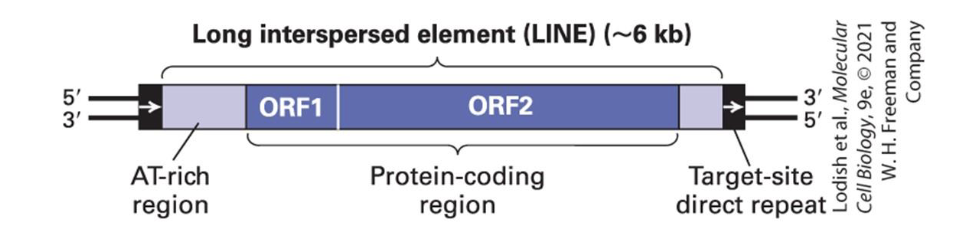
LINEs
900,000 of these in our genome - represents abt 21% of it
these are the most common retrotransposons
contain two open reading frames
ORF1
for RNA binding
ORF2
encodes a reverse transcriptase and a nuclease, mediating the insertion
SINEs
abt 300bp in length (much shorter than LINEs)
occur at 1.6million sites in our genome - accounting for abt 13% of it
do not contain ORFs
insert themselves in the genome similarly to LINEs w AT rich regions but are parasites of LINEs because use those regions to insert themselves into the region but do not code proteins
Alu
the most common SINE — aka most common repeated sequence in the genome
probably evolved from a non-coding RNA gene
how does the DNA insertion of LINEs work? (5 steps)
RNA is produced and exported from the nucleus
ORF1 (and RNA-bonding protein) and ORF2 (Reverse transcriptase and nuclease) are translated and bind LINEs’ RNA
RNA-protein complex imported to the nucleus
Nuclease cuts DNA at an AT-rich sequence and uses the DNA ends as primers for the reverse transcriptase of the LINE RNA
No transposase or integrase used
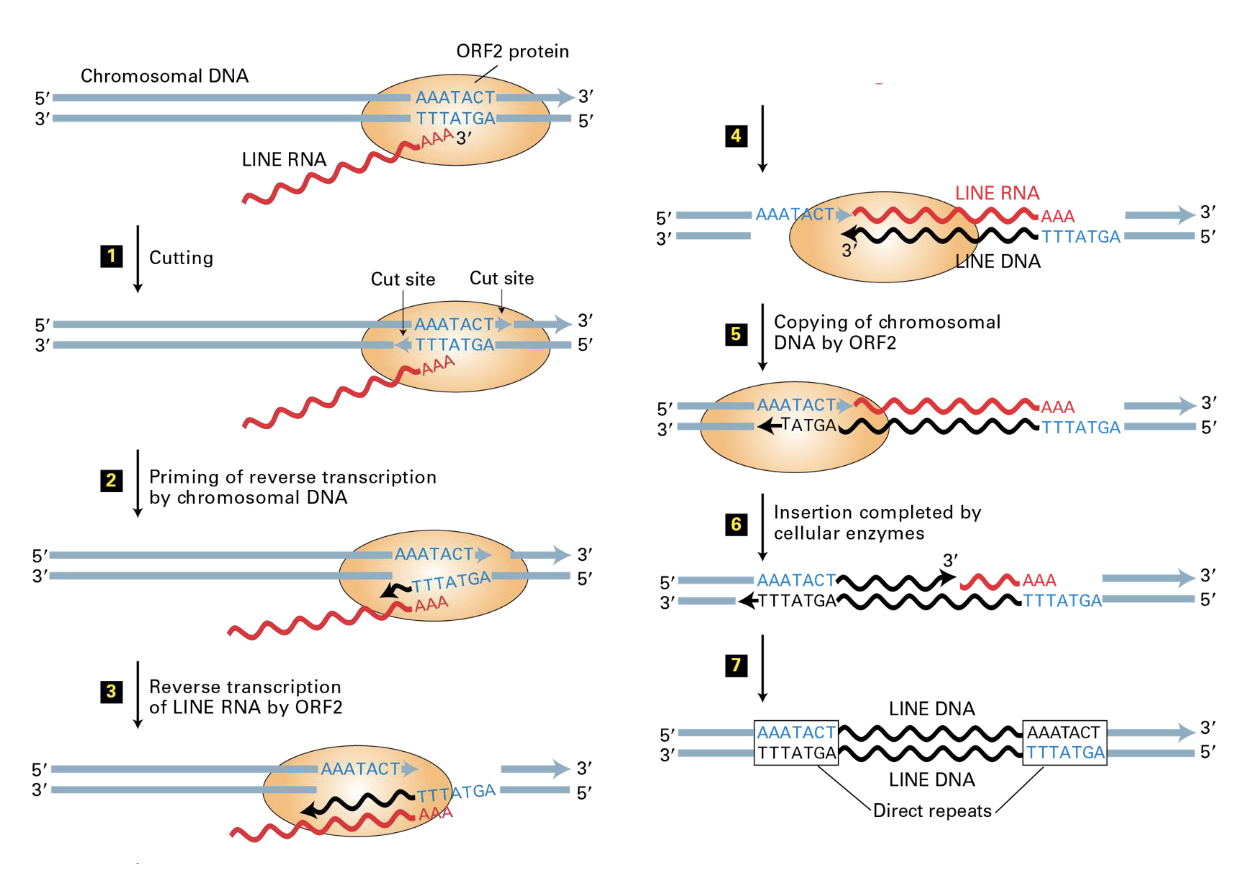
how do SINEs insert themselves?
they use ORF1 and ORF2
how do TE mvmnts lead to genome changes?
Recombination between repeated elements can shuffle exons and produce new genes with new combination of existing exons
helpful because can do recombination with non-homologous chromosomes
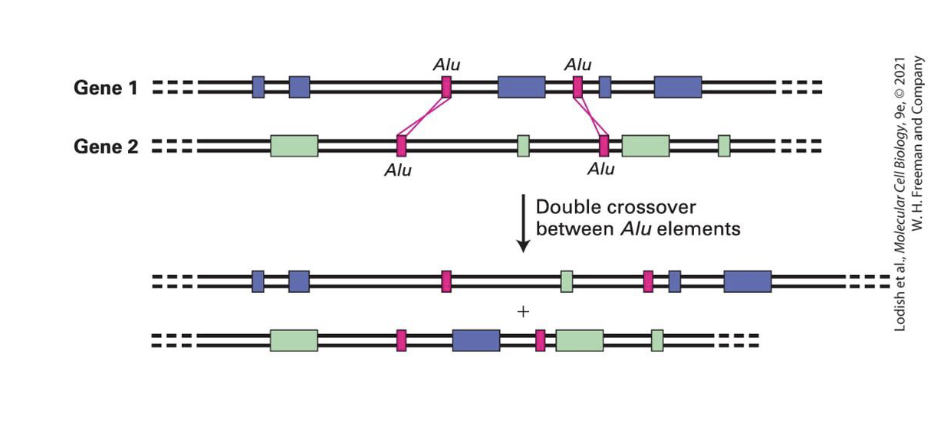
What can transposons and LINEs carry with them when they move that causes shuffling?
they can carry unrelated flaking sequences