T1 (copy)
1/77
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
78 Terms
1. monosaccharide
2. disaccharide
3. polysaccharide
1. mono: individual sugar monomer
2. di: molecules formed by condensation of 2 mono --> glycosidic bonds
3. poly: polymers formed by condensation of many mono- (repeating mono- unit)
explain how monosaccharides join to form dis. or polys.
through condensation reactions and forming glycosidic bonds
oligosaccharides
poysaccharides w/ 3-10 sugar units
saccharides levels

hexose glucose
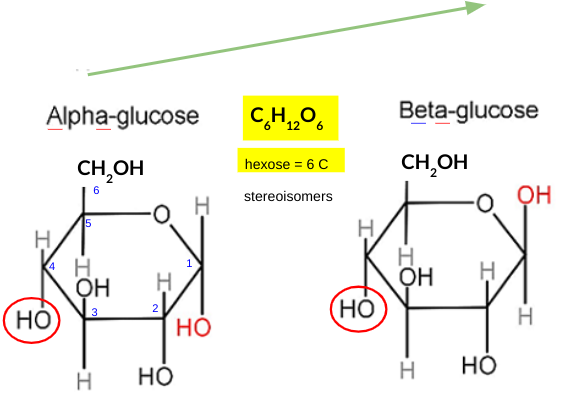
explain how the structure of GLUCOSE is related to its function
six-carbon sugar: high energy content, easily metabolized through glycolysis
hydroxyl groups: make it highly soluble in water, facilitating its transport in the bloodstream to various cells in the body
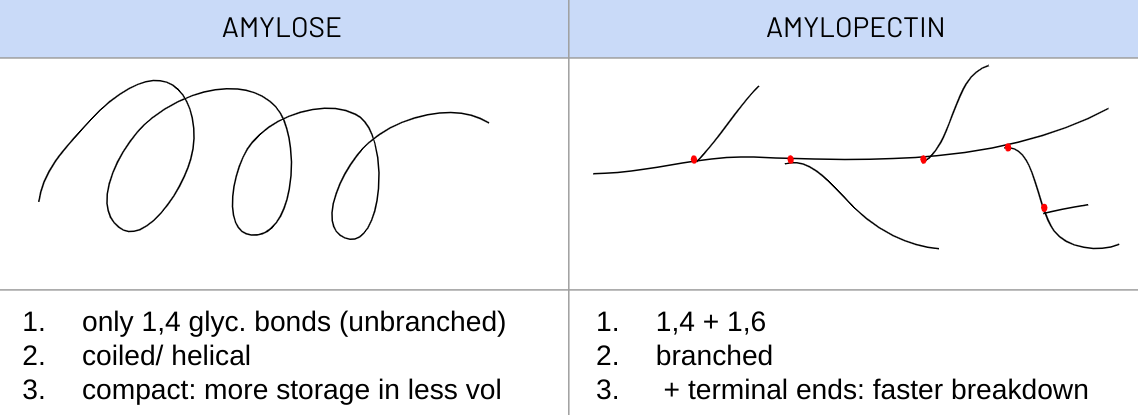
explain how the structure of STARCH is related to its funciton
F: energy storage - plants
· mixture of amylose + amylopectin - α glucose poly.
1. large molecules --> too large to diffuse out of cell
2. no distinct polarity --> unreactive
3. insoluble --> no osmotic effect
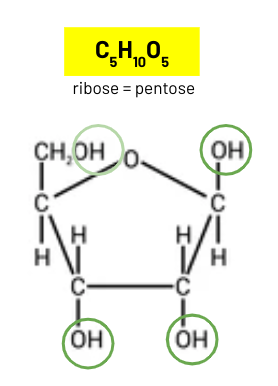
ribose
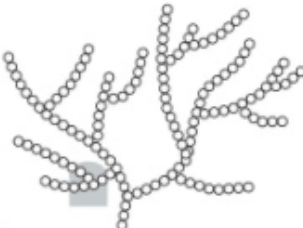
explain how GLYCOGEN structure related to its functio
F: energy storage - animals + fungi
· 1,4 + 1,6 glyc bonds --> α glucose monomers
1. compact: +in - vol
2. unreactive: no polarity
3. low solubility: no osmotic effect
4. large ≠ diffuse out of cell
5. more branches, more terminal glucose molecules, hydrolysed faster and supply of energy
animals = more active
types of glycosidic bond
1,4: results in linear polymer
1,6: results in branched polymer
glycosidic bond
between OH group + H
glycosidic bonds asily broken down
rapid release of glucose (mono) for cellular respiration
1,4 - non branched
1,6 - branched
structure cellulose
contain only beta glucose
unbranched, linear chains
alternate monomers rotated through 180 º
contain only 1,4 glycosidic bonds
cellulose molecules are bonded to each other by H bonds
polysaccharide of beta glucose
every other glucose is inverted
cellulose molecules arranged parallel/ as microfibrils
joined by H bonds
how structure of CELLULOSE relates to its function
plant cell walls
1. cellulose molecules are straight
2. many H bonds hold molecules/ chains together (microfibrils)
3. results: strong to prevent cell lysis, matiain turgidity, resist turgor pressure
4. polar nature of glucose allows water molecules to diffuse through
condensation reaction
water molecule removed from rectants and bond fromed
covalent bonds formed
· protein: peptide bond
· lipids: ester bond
· carbohydrates: glycosidic bond
hydrolysis
water molecule used + bond broken
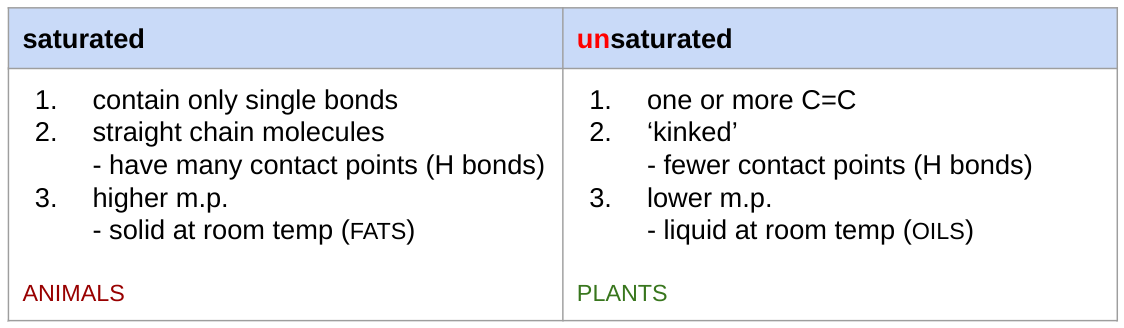
properties of saturated vs unsaturated fatty acids
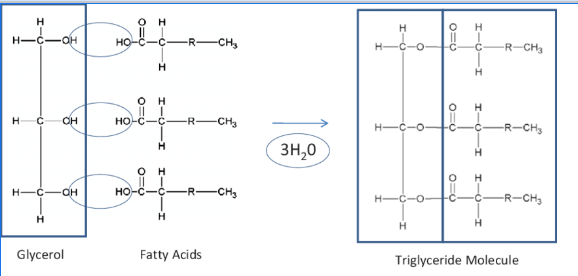
tryglyceride formation
(1) CONDENSATION reaction
(2) 1 glycerol + 3 fatty acids
(3) hydroxyl group + carboxyl group
(4) ester bond
(5) + 3 H2O
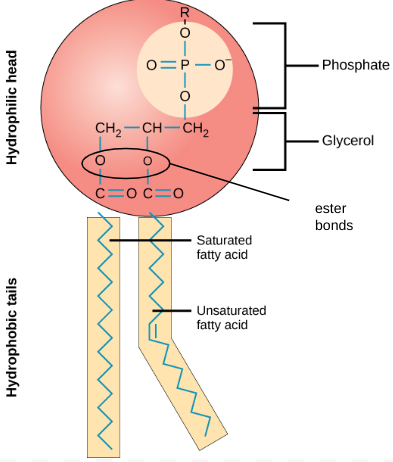
relate the structure of lipids to their function
1. high energy: to mass ratio
energy storage, high calorific value from oxidation
2. insoluble - non polar
no osmotic effect
used for water proofing - hydrophobic fatty
3. thermal insluation
slow conductor of heat
4. buoyancy
less dense than water
describe structure & function of phospholipids
amphipathc
glycerol backbone
- attached to 2 hydrophobic FA tails
- & 1 hydrophilic -vely charged phosphate head (PO3)⁻
= POLAR molecule
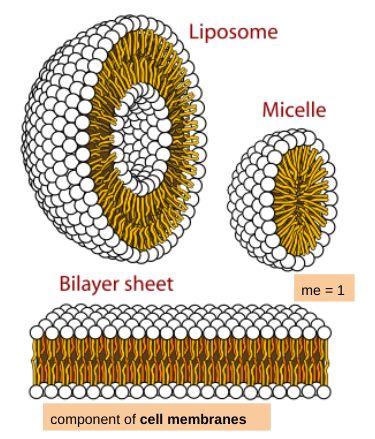
F: forms phospholipid bilayer: component of membranes
+ tails can splay outwards: waterproofing
phospholipid structures
structure amino acid
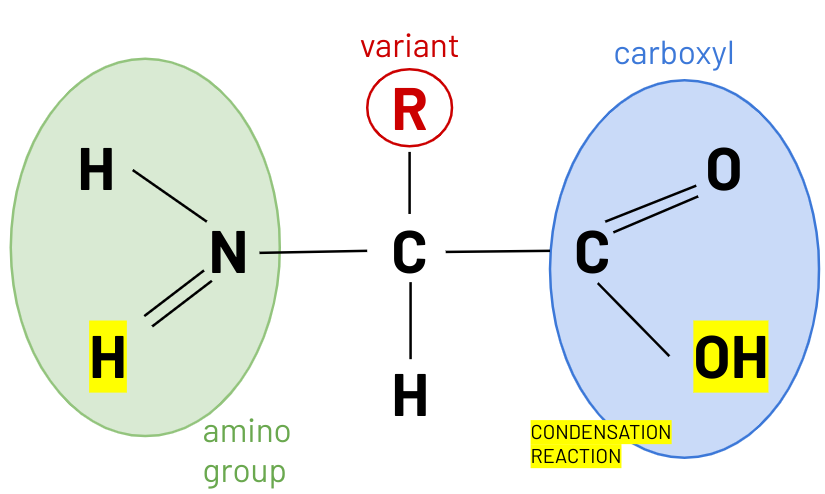
peptide bond
H amino group + OH carboxyl group
amino acid monomers --> form di peptides --> polypeptides
linked by peptide bonds formed in condensation reactions
primary structure of protein
the exact sequence and number of amino acids in polypeptide chains
determined by sequence of codons on mRNA
+ only peptide bonds
secondary structure of protein
a polypeptide chain folding
into alpha helix or beta pleated sheet
due to H bonds between different regions of polypeptide chains
H bonds between C(O)OH & NH2
describe 2 types of secondary protein structure
alpha helix:
all NH bonds on same side of protein shape
H bonds parallel to helical axis
beta pleated sheet:
NH and C=O groups alternate sides
tertiary structure of protein
3D further folding of proteins - supercoiling
complex 3D structures
held together by bonds between R groups of AA along chain
1. hydrogen bonds (numerous and easily broken)
2. ionic bonds (2nd strongest, between charged R groups, change pH interrupts them)
3. disulphide bridges (strong covalent S-S) - CYSTINES
quarternary structure of protein
more than 1 polypep chain
same bonding as 3tiary
- between R groups of diff chains
can have a prosthetic group - non-protein compontnet
--> result in conjugated molecules
explain how the sequence of amino acids determines the shape/properties of a protein
primary structure: seq of aa that determines 3tiary structure
because aa/ R groups determine position bonds (hydrogen, disulphide, ionic)
polar aa/ R groups need to be on outside so it can dissolve into plasma
final structure of molecule has to be specific shape to be complementary/bind to the receptor molecules
or have active site
describe funtion and structure of globular proteins
· spherical and compact
· usually (semi) water soluble - form colloids in water
· involved in metabolic processes eg. enzymes, heamoglobin, hormones
· complex 3tiary/ 4arternay structures held together by (our 3) bonds
explain why globular proteins are soluble in water (4)
protein folded so hydrophilic R groups face outwards, hydrophobic R grps in
these exposed R groups are charged: polar / ionic
therefore they form H bonds with water
because water is a polar solvent / dipole nature
describe funtion and structure of fibrous proteins
· form long parallel chains/ fibres
· very little or no 3/4 structure - mainly 2dary
· occasional cross-linkages which form microfibriles for tensile strength
· sequences of aa repeat
· insoluble in water
· useful for structure and support. eg: collagen
function collagen
structure: component of bones, cartilage, connective tissue, tendons
structure collagen
fibrous protein:
· 3 polypeptide chains
· that form a triple helix
· helices held toegether by hydrogen bonds
· many tropocollagen molecules (triple helices) joined together
· form long parallel chains/ fibres
· very little 3/4 structure - mainly 2dary
· STABLE alpha triple gamma helix - repeating aa seq: glycine-proline-other
· insoluble in water
· cross-linkages: H bonds and staggered covalent bonds between fibres form microfibriles and = high tensile strength
explain significance of repeating amino acid sequences in formation of tropocollagen
glycine is very small so collagen fibres close together
R group = single H
allows formation of hydrogen bonds to hold polypep. chains together
funtion haemoglobin
binds to O2 with variable affinity to transport it around the body in bloodstream
structure heamoglobin
globular protein:
· spherical and compact
· 2 alpha and 2 beta chains, 4 prosthetic haem groups: conjugated
· hydrophilic R groups face outwards, hydrophobic R grps in
--> water soluble - dissolves in plasma
· Fe2+ haem groups forms dative bond with O2
· 3tiary structure changes so it is easier for subsequent O2 molecules to bind
role in plants of 4 inorganic ions
1. phosphate ions - to make ADP & ATP
2. magnesium ions - produce chlorophyll
3. nitrate ions - DNA and amino acids
4. calcium ions - to form calcium pectate for middle lamella
calcium pectate
sticks cell walls together
found in middle lamella
explain how dipolar nature of H2O is essential dor living organisms
hygrogen bonds, polar
1. water is a polar solvent - used for transport medium
2. has high surface tension - allows pond-skaters
3. high specific heat cap. - thermoregulation
4. water max dense at 4ºC
5. incompressible - used for hydraulics & structural support (turgor changes)
nucleic acids
polymers of nucleotides
RNA (ribonucleic acid) & DNA (deoxyribonucleic acid)
structure of nucleotides
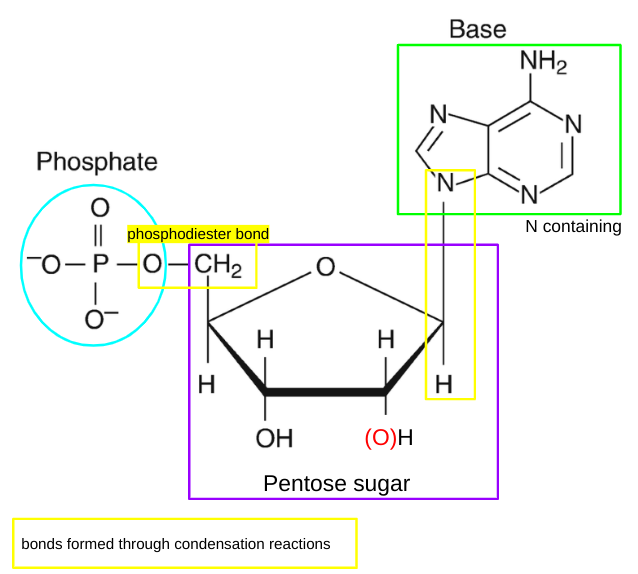
nucleotides definition
individual monomers that make up polynucleotides which are compsed of a phosphate group, a pentose sugar and a nitrogen containing base
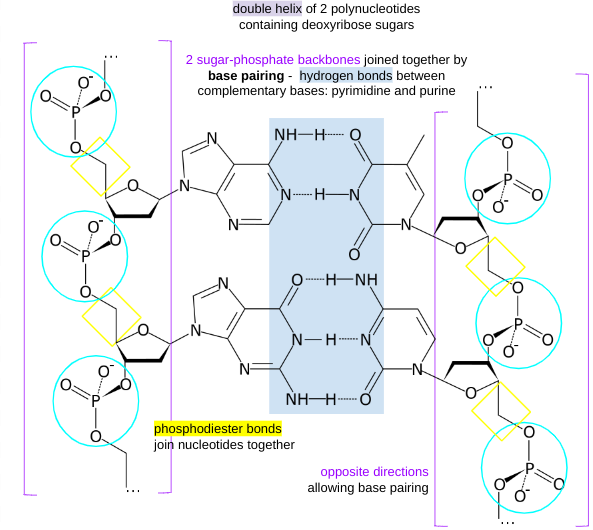
structure of DNA
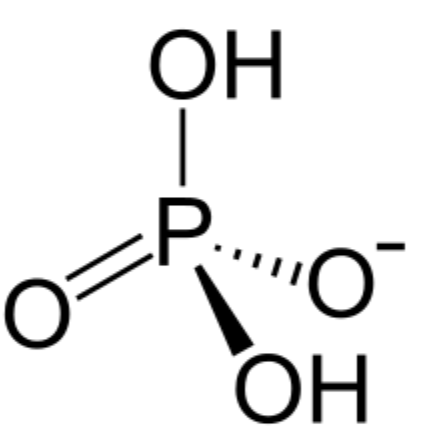
phosphodiester bonds
two of the hydroxyl groups in phosphoric acid react with hydroxyl groups on other molecules to form two ester bonds
found in DNA and RNA backbone
2 in phosphodiester bonds, 1 loses H to gain -1 charge
pentose sugar in nucleotide in DNA and RNA
deoxyribose
ribose
organic bases in DNA
Adenine
Cystosine
Guanine
Thymine
purine bases
have 2 nitrogen containing rings
Adenine and Guanine
organic bases in RNA
Adenine
Cystosine
Guanine
Uracil
pyramidines
contian 1 nitrogen containing ring, single ring structure
pyramidines - Thymine, Cystosine, Uracil
ribose and deoxyribose structure
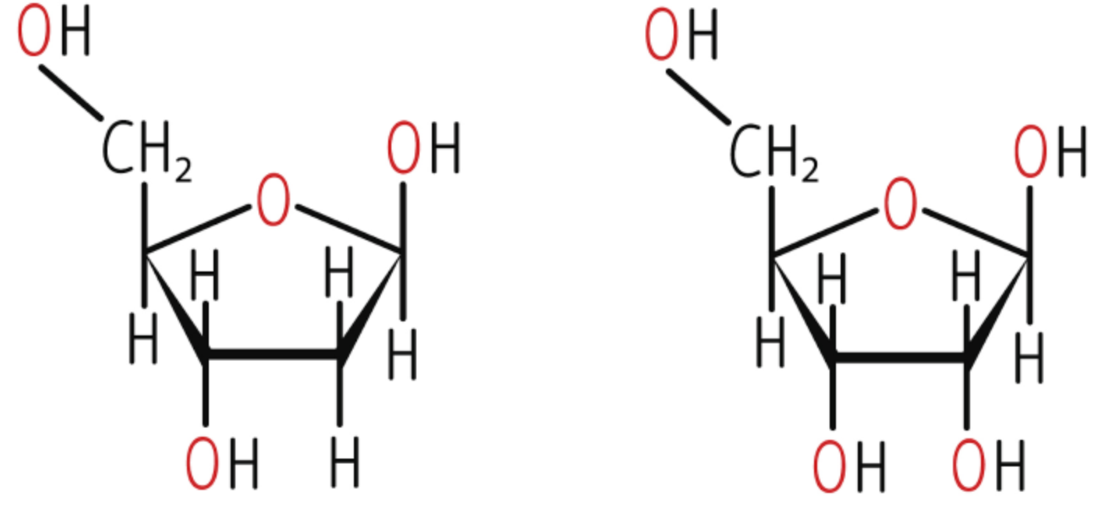
base pairing
A-T: 2 H bonds
C-G: · H bonds
RNA
single stranded
can form complex shapes through H bonds
comes in multiple different forms
made of nucleotides (AGCU)
mRNA - messenger: sugar phospate backbone
tRNA - transfer
rRNA - ribosomal
which are involved in protein synthesis
DNA replication
semiconservative
1. DNA double helix unwinds forming a replication fork
· H bonds compl. bases broken
· catalyst: DNA helicase
2. both free strands used as templates
· free nucleotides line up
· complementary base pairing occurs between template & free nucleotides
3. adjacent nuclotides (which contain the bases) joined
· condensation reaction
· phosphodiester bonds
· catalyst: DNA polymerase
----------------------------------------------------------------------
DNA polymerase only works in 5' to 3' direction
strands run in opposite directions
leading strand: in 5' --> 3' direction: made continuously by DNA polymerase
lagging strand: in 3' --> 3' direction
made as a series of small chunks at a time in 5' to 3' direction between primers forming okazaki fragments
ligase seals fragments in both strands w ph.diester bonds
----------------------------------------------------------------------
4. 2 identical new DNA molecules automatically fold: double helices
· H bonds within molecules
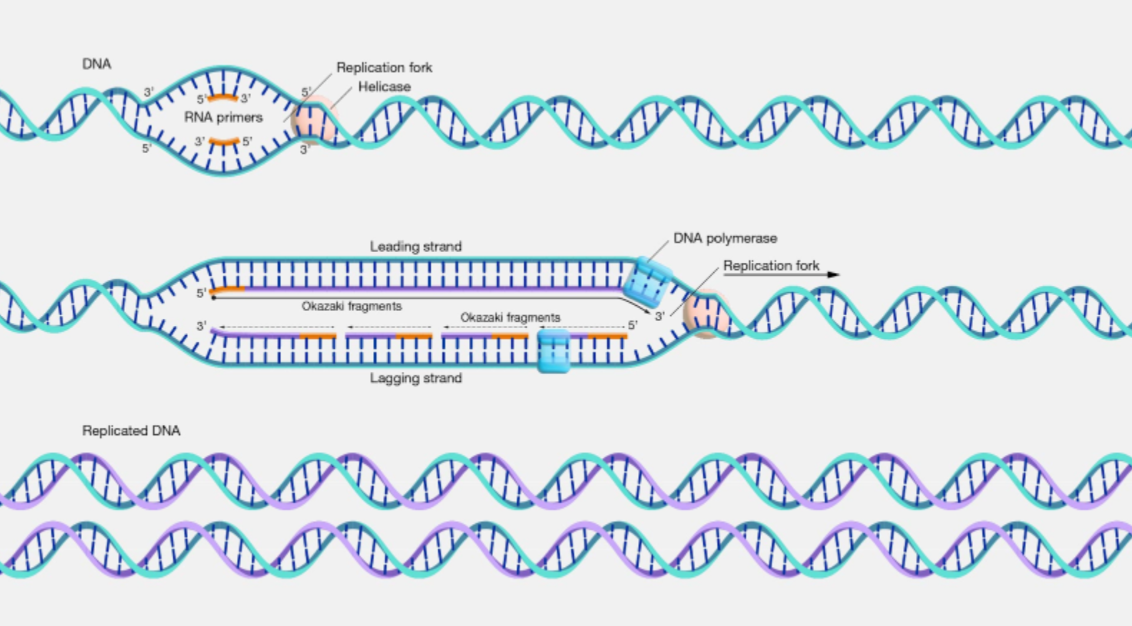
enzymes involved in DNA replication and function
DNA helicase - unwinds DNA helix by breaking H bonds between bases
DNA polymerase - catalyses formation of phosphodiester bonds between nucleotides during the synth of a new DNA strand
ligase - enzyme which joins Okazaki fragments on lagging strand together or any other fragments by forming phosphodiester bonds between them
semi-conservative replication definition
new DNA molecules contain 1 original strand and 1 newly synthesised strand
· ensures genetic continuity between generations of cells
· genetic info passed from one gen to the next
gene definition
sequence of bases on a DNA molecule coding for a sequence of amino acids in a polypeptide chain
(explain) nature of genetic code
· consist of triplets of bases: codons, which each code for 1 amino acid
· contains (1) start and (3) stop codons: maks start/stop protein synth, establish reading frame
· degenerate: more than one triplet codes for the same amino acid
this reduces the effect of mutations
· non-overlapping: each triplet only read once, triplets don't share bases
· not all of the genome codes for proteins: non-coding regions (introns)
coding regions = exons
· universal: same in all organisms + species
degenerate nature of genetic code effect
mutations have less effect:
more triplet codes than amino acids
some mutations have no effect on protein made as new triplet may still code for same amino acid
helps mantain same structure + function of protein
deletion/ insertion: more likely harmful, causes ‘frameshift’
all codons ‘downstream’ of the mutation are read differently
gene mutations
changes to base sequence:
deletions: nucleotides not incorporated into chain
insertions: extra nucleotide(s) incorporated into growing DNA chain
substitutions: incorrect nucleotide incorporated into chain
frame shift: (1st two)
more harmful
all codons downstream of mutation are read diff and likely produce diff aa
start and stop codons
start: TAC
stop: ATT, ATC, ACT
enzymes def
biological catalysts
bio: globular proteins: synthesised by ribosomes from mRNA.
catalysts, lower activation energy: minimum amount of energy particles must collide with in order to react.
control RoR of metabolic R
anabolic: build new chemicals
catabolic: break subs down
place of actions of enzymes: intra and extracellularly
how do enzymes lower activation energy
form enzyme/substrate complex:
active site affects bonds in substrate, less energy req to break them
reacting substances are brought close together, easier for bonds to form between them
once ration complete, product no longer fits active site and leaves
induced fit hypothesis
active site: still has specific shape but is flexible
substrate enters active site
AS undergoes small conformational changes to fit substrate better
/ shape of active site is modified around it to form the active complex
this puts strain on substrate bonds, lowering activation energy
once products left complex, enzyme reverts to inactive form
specificity of enzymes
result of very specific active site shapes arising from seqs of aa folded in a particular way: 1/2/3/4 structure/ folding
.: each enzyme only compl to one type of substrate and will only cat that reaction
types of inhibition of enzyme activity
reversible: enzyme not permanently damaged. once inhibitor removed, enzyme activity functions normally
competitive inhibition
non-competitive inhibition
irreversible:
inhibitor combines w enzyme by permanent covalent bonding, impeding catalysis
never “natural”: not used as a means to control metabolism
slower, but much more damaging effects
end-product inhibition
how enzymes are affected by competitive inhibition
inhibitor molecule similar in shape to substrate molecule, binds to active site, forms enzyme-inhibitor complex
temporarily prevents ES complexes forming until released, decr RoR
competes w substrate for binding at active sites: the more inhibitor mols there are, less likely substrate will bind to enzyme
level of inhibition depends on levels of substrate and inhibitor (game of probability/ numbers)
how enzymes are affected by non-competitive inhibition
inhibitor binds to enzyme/ forms complex w enzyme or ES complex
→ forms bonds w enzyme at allosteric site
trigger conformational change/ change of shape of active site
→ prevents substrate binding, can no longer catalyse reaction
not on the active site: inhibitor not competing for the active site
→ only level of inhibitor affects level of inhibition. concentration of substrate does not affect level of inhibition
how enzymes are affected by end-product inhibition
an (end) products of the reaction (pathway) acts as competitive or non-comp inhibitor for an enzyme involved in (beginning of) pathway
prevents further formation of products: feedback control
—> if levels high: inhibits formation of more product, if low, no enzyme inhibition
regulatory enzymes: can have molecules can bind to them and bring about non-competitive inhibition (not active site)
how to compare effect of an enzyme
measure initial RoR
bc RoR decreases over time, initial is fastest rate
bc as R progresses substrate is used up/ broken down, conc decrs
so less substrate can collide w enzyme + form complexes
initial: substrate is in excess, not limiting, subs conc should not limit RoR
subs conc is no longer controlled
before end prodcut inhibition can occur
how to measure initial RoR
calc initial rate: tangent to graph at t=0
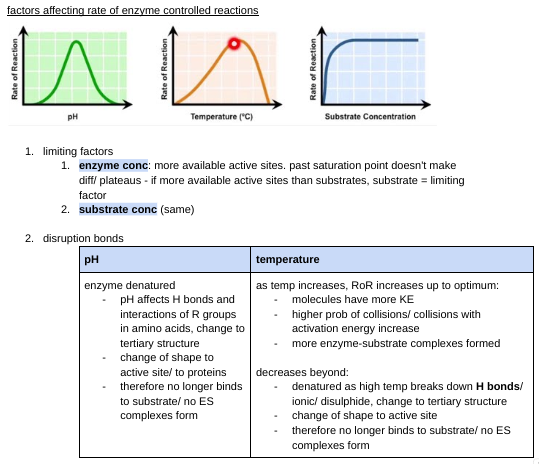
factors affecting rate of enzyme controlled reactions
stages of protein synthesis
transcription
mRNA leaves thru nuclear pores, travels to ribosomes/ RER
translation (protein synth)
modified (4º structure produced) and packaged into vesicles: golgi apparatus
exocytosis releases protein
Describe the process of transcription (4)
in nucleus
DNA strands separate
antisense strand used as template for mRNA
mRNA forms on antisense strand in order to code for a peptide
coding strand of DNA= sense strand
is identical to coding strand of DNA
RNA polymerase binds to open DNA and synthesizes mRNA
tRNA structure
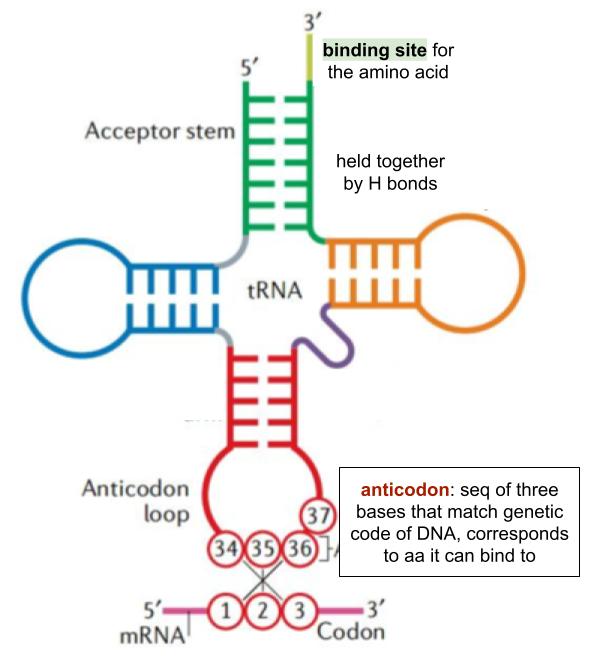
role of ribosomes in protein synthesis
translation
to hold the tRNA on mRNA
whilst peptide bonds form to join adj amino acids together
describe the process of translation
in cytoplasm/RER, in ribosomes
mRNA attached to ribosome
tRNA is attached to a specific amino acid, brings aa
tRNA anticodon binds to mRNA codon. compl ase pairing (H) ➰ tRNA +mRNA
peptide bond form ➰ amino acids
process involves start/ stop codons
Describe how monomers are bonded to a polypeptide chain during the synthesis of a protein
formation of peptide bond
between amino acids and carboxyl gr
by condensation R
effect of point mutations on amino acid sequences:
changes triplet/ codon, may change amino acid coded for
if affects protein produced, can change phenotype
eg. sickle cell anemia: gen disease, produces sickle shaped RBCs due to point mutation that causes a change in a single amino acid in the polypeptide sequence
hemoglobin formed forms rods, gives RBC that shape
oxygen not carried efficiently
cannot reach narrow blood vessels