Part 2 108A Final (chapter 11, 25, 26, 27)
1/91
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
92 Terms
Integral membrane proteins
Span the lipid bilayer and interact with the hydrophobic core.
They are often alpha-helical or beta barrel structures.
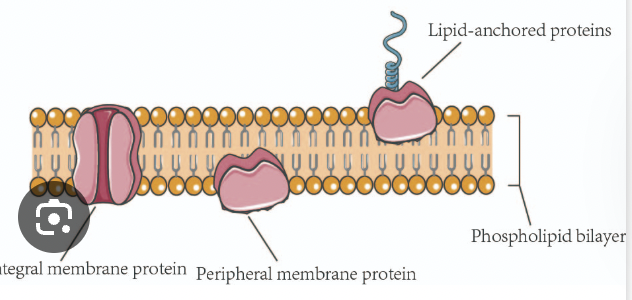
Peripheral membrane proteins
Attach to membrane surface via non-covalent interactions or to integral proteins
Do NOT interact with hydrophobic core
These are on the outside
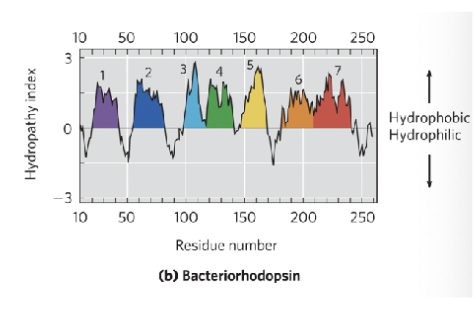
Hydropathy plot
Graph of hydrophobicity vs amino acid position; predicts membrane spanning segment
X axis represents the amino acid sequences
Y axis represents hydrophobicity (positive values indicate hydrophobic regions and negative values indicate hydrophilic regions)
By analyzing peaks and valleys, you can identify potential membrane spanning domains (such as if the graph shows a region with hydrophobic and hydrophilic areas we can interpret this as a possible transmembrane protein)
Membrane fusion
Process of two lipid bilayers merging to become one
Two separate bilayers merge into a continuous bilayer
Ex: Fusion of synaptic vesicles with the cell membrane in nerve cells release neurotransmitters to transmit signals or viral entry into host cells
v-SNARE
Proteins on the vesicle that mediate vesicle fusion
ON the vesicle membrane
Cytoplasmic face of neurotransmitter vesicle
t-SNARE
Proteins on the vesicle that mediate vesicle fusion
ON the target molecule
Contained in target molecule (located on the target molecule)
What do v-SNARE and t-SNARE do?
These proteins interact to form a stable trans-SNARE complex, bringing the vesicles and target membrane close together and leading to membrane fusion
Facilitated diffusion
Passive transport via membrane proteins (channels or carriers, NO ATP)
SERCA Pump
A P-type ATPase that pumps Ca2+ sarcoplasmic reticulum using ATP
its primary function is to transport Ca2+ ions from the cytoplasm into the sarcoplasmic reticulum (SR) or endoplasmic reticulum (ER) using energy from ATP hydrolysis
This helps maintain low Calcium concentrations in the cytoplasm, which is essential for various cellular processes
V-Type ATPases
V type stands for vacuoles that hydrolyze energy (ATP) to create ion gradients (vicky and licky make energy gradients)
Vacuolar
Pumps protons into organelles (lysosomes) using ATP
Located in various intracellular membranes like lysosomes and vacuoles and they primarily hydrolyze ATP to create ion gradients for cellular processes
F Type ATPase
F type, First type best type, found in mito and chloro - obvi best parts of the cell, synthesizing the energy of the cell
They are primarily found in mitochondria and chloroplasts, where they synthesize ATP using electrochemical proton gradients generated by respiration or photosynthesis
Na+/glucose transporter (sodium and glucose transporter) (SGLT)
A protein that uses the electrochemical gradient of sodium ions to transport glucose across cell membranes
Secondary active transport (does not directly use ATP but relies on the movement of Na+ down their concentration gradient to power the movement of glucose against its concentration gradient)
Approximate value of proteins
30%-70%
Vary based on specific membranes function and location within the cell
Example: The inner mitochondrial membrane involved in ATP production is about 75% protein by mass while the myelin membrane contains less than 25% protein.
Approximate value of Phospholipids
7% to 40%
Structural component of cell membranes
Approximate value of Sterols (cholesterol)
0%-25%
Cholesterol is the major sterol in animal cell membranes comprising about 30% of the lipid bilayer on average
What are photoreceptor disc membranes characterized by?
A high concentration of rhodopsin (over 90% of the membrane protein)
Lipid-Anchored protein
Covalently attached to lipid molecules embedded in the membrane
What are two common features about the amino acid composition of transmembrane proteins?
Abundance of hydrophobic residues (Leu, Val, Ile) in the transmembrane region - crucial for their integration and stability within the lipid bilayer which is a hydrophobic environment formed by the fatty acid tails of membrane lipids
Tyr and Trp residues and lipid water interface- their amphipathic nature helps anchor the protein at the boundary between the hydrophobic membrane interior and aqueous environment
How many Amino acids span the membrane in an alpha helix?
Around 20 non-polar amino acids span around 30 angstroms hydrophobic core of a bilayer
An alpha helix rises 1.5 A per residue, so 30A/1.5A = 20 residues
Each alpha helical turn is 3.6 residues, spanning the membrane takes about 5-6 turns
Transcriptome
The complete set of RNA transcripts (including mRNA, tRNA, rRNA, and other non-coding RNAs)
What does it mean that membrane lipids are amphipathic?
They have both hydrophilic (polar) head groups and hydrophobic (nonpolar) fatty acid tails
Why do lipids spontaneously assemble in water.
To minimize free energy by segregating hydrophilic heads toward water and hydrophobic tails away from water
What is a micelle and when is it formed
A spherical lipid structure formed by single-tailed lipids (detergents) with hydrophobic tails inward and polar heads outward
What is a lipid bilayer
Two leaflets of lipids forming a planar or curved sheet that encloses an aqueous compartment (vesicles or lipsomes)
Approximately how thick is a lipid bilayer?
About 3 nm (30 A) thick
Are the inner and outer leaflets of a lipid bilayer identical
No they are asymmetrical meaning they have different lipid compositions
What does membrane fluidity mean?
Lipids and proteins can move laterally within the membrane, contributing to its dynamic nature.
What is the functional purpose of the lipid bilayer’s barrier?
It provides selective permeability, separating the cell from its environment and maintaining distinct internal compartments
What are the key criteria that must be met for membrane fusion to occur
Membrane continuity must be preserved
Membranes must recognize each other
Close proximity with water removal (local dehydration)
Fusion proteins (eg SNAREs) must mediate the process
What structural changes happen during membrane fusion?
Increase in local membrane curvature
Disruption of bilayer at contact point
Hemifusion (outer leaflets fuse first)Full fusion creates a single continuous bilayer
What roles do v-SNAREs and t-SNAREs play in membrane fusion?
They interact to bring membranes close together, overcome energy barriers, and facilitate bilayer distortion for fusion
What additional requirements are often needed for membrane fusion during endocytosis or secretion?
A trigger or signal (calcium or ligand binding) and energy input (ATP or GTP hydrolysis)
What type of transport is Glut1?
Facilitated diffusion of glucose; passive transport follows the concentration gradient
Transports glucose into RBCs
Specific for glucose over sugars
Glut1 is a type III integral membrane protein, meaning it spans the cell membrane multiple times with both its N and C termini inside the cell
How would a membrane adjust to become more liquid-disordered until certain temperature changes (more fluid)
Increase unsaturated fatty acids (kinks/double bonds)
Shorter fatty acid chains
Higher temperature
How would a membrane adjust to become more liquid-ordered until certain temperature changes (less fluid)
Increase cholesterol
More saturated fatty acids
Lower temperature
What is the equation for delta G of transport for an uncharged solute?
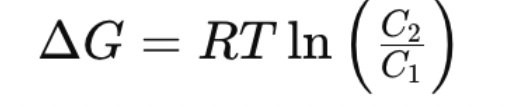
What is the equation for delta G of transport for an charged solute?
Same equation for an uncharged solute plus Z (charge) times F (faradays) times membrane potential
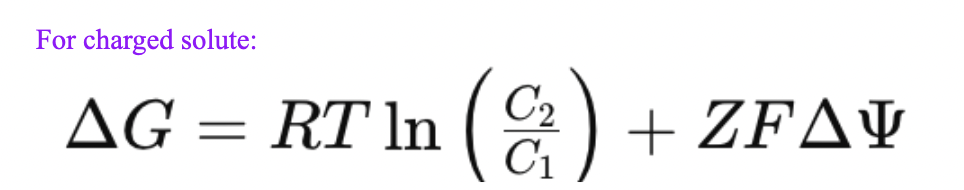
What is the primary function of P-type ATPases
To actively transport cations (Ca2+, Na+) against their concentration gradients using ATP through reversible phosphorylation of an aspartate residue
What are the three cytoplasmic domains of P-type ATPase and their functions?
N Domain: Binds ATP and Mg2+, phosphorylates Asp in P domain
P Domain: contains the Asp residue that gets phosphorylated
A domain: acts as activator, removes the phosphate to reset the cycle
How does the mechanism of P-type ATPases drive ion transport?
ATP binds to the N domain
Phosphate is transferred to Asp in the P domain
Conformation change (E1—> E2) moves the ion across membrane
A domain removes phosphate, resetting the protein
What is the SERCA pump and how does it function
A P-type ATPase in the sarcoplasmic reticulum of muscle cells
Pumps 2 Ca2+ ions into the SR lumen per ATP hydrolyzed
Phosphorylation shifts it from E1 (high Ca2+ affinity) to E2 (low Ca2+ affinity)
Essential for muscle relaxation
Okazaki Fragment
Short DNA segments (1000-2000nt in bacteria, 150-200 nt in eukaryotes) synthesized discontinuously on the lagging strand during DNA replication. Later joined by DNA ligase
DNA Pol I
Not the main polymerase for replication. It has low processivity and functions mainly in the removal and replacement of RNA primers, as well as in repair and cleanup
DNA Poly III - repliaction
The main replicative polymerase in E.Coli. It adds nucleotides during elongation on both the leading and lagging strands.
RNaseH1
Type of replication enzyme
-Specialized nuclease that degrades RNA in RNA-DNA hybdris
Not typical, in case something goes wrong
Topoisomerase
An enzyme that relieves topological stress ahead of replication forks by cutting and rejoining DNA strands. Topoizomerase IV resolves catenanes during termination
replication
Ligase
Seals nicks in the DNA backbone after primers are replaced, forming a continuous strand
Primase
Enzyme that synthesizes short RNA primers (10-60 nt) to initiative DNA synthesis in E.Coli, it's known as DnaG, and works with helicase.
Catenane
Two interlinked circular DNA molecules formed after replication of circular chromosomes. Separated by topoisomerase IV.
What does bidirectional replication mean?
Replication proceeds in both directions from a single origin (oriC in E.Coli) creating two replication forks moving away from the origin
How does DNA proofreading occur?
DNA polymerase (like DNA pol III) have 3’--> 5’ exonuclease activity
If the wrong nucleotide is added it alters the geometry
The polymerase pauses and removes the incorrect base
This improves accuracy 100-1000 fold
α (alpha)
Polymerase activity
allen loves pollys ass
β (beta)
Sliding clamp for processivity
You beta slide that clamp over
ε (epsilon)
3’ to 5’ exonuclease (proofreading)
looks like an e for epsilon or exonuclease (always works 3—>5)
θ (theta)
stabilizes ε, part of core polymerase
stabilizes the exonuclease
Theres an e in theta, its meant to be
theta and episolon love eachother and theta wants to stabilize epsilon and become part of the core polymerase
Stabilizes the exonucease
τ (tau)
links multiple core polymerases
Alpha Taus link with multiple people
γ, δ, δ′
clamp loader complex
gamma delta delta primes have a clamp loading complex
meaning they really like to clamp
χ, ψ
bind to clamp loader
The ψ (psi) subunit joins the minimal complex by interacting with one of the γ subunits, forming a six-subunit complex (γ3δδ′ψ).
The χ (chi) subunit then binds to the ψ subunit to form the seven-subunit clamp loader (γ3δδ′χψ).
Chi and psi bind to the clamp loader to make it a 7 subunit clamp load
Chi Psis love a good seven man
RNA polymerase
Enzyme that synthesizes RNA from a DNA template. It adds ribonucleotides to the 3’-OH end in the 5’ to 3’ direction using ATP, UTP, and CTP and requires Mg2+
rho-dependent termination
In E.Coli this uses the rho protein, an ATP-depedent RNA-DNA helicase. Rho binds to CA rich sequence (rut site) and moves along the RNA to unwind the RNA-DNA duplex and release the transcrit
rho-independent termination:
Occurs via a self complementary region in the RNA that forms a hairpin loop followed by a string of U residues. This structure causes RNA polymerase to pause and release the RNA.
Intron
A non coding sequence in a eukaryotic primary mRNA transcrip that is removed during splicing
Exon
A coding region of an mRNA transcript that remains after splicing and is translated into protein
TATA box
A promoter sequences (TATA(A/T)A(A/T)(A/G) found 30 bp upstream of the transcription start site in many Pol II genes. It helps position RNA polymerase for initiation
DNA Pol II
responsible for the synthesis of mRNAs and many ncRNAs
What are the correct order of events for transcription?
Initiation- RNA polymerase binds to promoter (TATA box)
Promoter clearance- enzyme begins RNA synthesis and escapes promoter
Elongation - RNA is synthesized in the 5’ to 3’ direction but adds bp 3—>5
Termination- RNA transcript is released and polymerase dissociates
Prokaryotic Initiation
RNA pol holoenzyme (core + σ factor) binds to promoter sequences (eg, -35 TTGACA, -10 TATAAT)
No primer is needed
σ70 helps recognize promoter, dissociates as elongation begins
Prokaryotic Elongation
RNA pol synthesizes RNA complementary to template strand
DNA unwinds, forming a transcription bubble
NusA replaces σ and aids elongation
Prokaryotic Termination
Rho-independent: hairpin + U-rich sequence causes release
Rho-dependent: Rho protein unwinds RNA-DNA duplex at rut site
Eukaryotic Initiation
Involves RNA polymerase II and transcription factors (TFs)
Common promoter motifs: TATA box (around -30), Inr (+1)
A pre-initiation complex forms with Pol II and TF’s
CTD (C-terminal domain) of Pol II is phosphorylated to begin elongation
Eukaryotic Elongation
RNA synthesizes in 5’ to 3’ direction
DNA reanneals behind the transcription bubble
Eukaryotic Termination
Pol II transcribes past the cleavage site
RNA is cleaved and Poly(A) tail is added
Polyadenylation signals, no Rho
5’ capping - mRNA processing
Adds 7-methylguanosine cap via 5’,5’ triphosphate linkage
Protects from degradation and helps ribosome binding
Splicing - mRNA processing
Introns are removed, Exons are joined
Carried out by spliceosome or ribozymes
3’ Polyadenylation in mRNA processing
Poly(A)tail (80-250 A’s) added to polyadenylate polymerase
Protects from degradation and aids in export and translation
This occurs in eukaryotic transcription because the new mRNA must be exported out to the cytoplasm, whereas the prokaryotic mRNA does not translation occurs simultaneously with transcription
RNA pol I
transcribes most rRNAs (eg 18S, 28S)
RNA Pol II
Transcribes mRNAs and many ncRNAs
works in eukaryotes
RNA Pol III
Transcribes tRNAs, 5S rRNA, and other small RNA’s
What are the steps in rho-dependent termination in E. coli?
Rho protein binds to the rut site (C-rich region on RNA)
It moves 5’ to 3’ using ATP hydrolysis
Catches up with RNA polymerase stalled at a termination site
Unwinds RNA-DNA hybrid → releases RNA transcript
Codon
A group of three nucleotides on an mRNA that encodes a specific amino acid
There are 64 codons (43 combinations), sufficient for encoding 20 amino acids
The genetic code is degenerate: multiple codons code for the same amino acid
There are 3 stop codons and 1 initiation codon (AUG)
Wobble hypothesis
Explain the ability of some tRNAs to pair with more than one codon due to flexibility at the third position of the codon
First base of anticodon:
A or C → specific pairing with U or G
U or G → wobble pairing: U pairs with A or G; G pairs with U or C
Inosine (I) in anticodon can pair A, U, or C
Only ~32 tRNAs are needed to read all codons due to wobble
aminoacyl-tRNA synthetases:
Enzymes that attach the correct amino acid due to its corresponding tRNA (charging)
Highly specific: usually 1 synthetase per amino acid
Two steps:
Formation to aminoacyl-AMP
Transfer to tRNA’s 2’- OH (Class I) or 3’-OH (Class II)
Some have proofreading ability, Ile-tRNA synthetase removes mischarged Val
EF-tu (Elongation Factor Tu)
Binds GTP and delivers aminoacyl-tRNA to the A site during elongation
After GTP hydrolysis, EF-Tu-GDP is released
IF-2
Binds GTP and helps position fMet-tRNA at the P site during initiation
Facilitates binding of the 50S subunit after correct anticodon-codon pairing
aminoacyl-tRNA
A tRNA molecule charged with its specific amino acid
The amino acid is linked via ester bond to the tRNA’s 3’ terminal A residue (on CCA tail)
Peptidyl transferase
Catalytic function of the ribosome (specifically the rRNA)
Forms peptide bonds between amino acids during elongation
Transfer peptide from tRNA in P site to the amino group of aminoacyl-tRNA in A site
No proteins are near the active site → Ribosome is a ribozyme
Bacterial ribosome big and small subunit
70S (30S + 50S)
Eukaryotic ribosome
80S (40S + 60S)
What is in ribosomes
rRNA and proteins —> 65% rRNA (catalytic core)
Ribosome is a ribozyme not a protein enzyme
What are the three binding sites in a Ribosome
A site = accepts aminoacyl-tRMA
P site = Holds tRNA with growing polypeptide
E site: Exit site for uncharged tRNA
Structure of tRNA
73-93 nucleotides in length
2D: Cloverleaf; 3D: L- shaped
Contains modified bases (methylated residues)
What are the key structural features and functions of tRNA in protein synthesis?
Size: 73–93 nucleotides, single-stranded RNA (ssRNA)
Structure:
5′ end: Guanylate (pG) residue
3′ end: CCA sequence for amino acid attachment
Anticodon arm: Contains anticodon (wobble at 5′ position of anticodon)
Cloverleaf shape (2D), twisted L shape (3D)
D arm & TC arm: Help with ribosome recognition
Function:
Adapter molecule: links specific amino acids to mRNA codons during translation
Ensures the correct amino acid is added to the growing polypeptide chain