Oxford Insight, Cooper "The Cell" Chapter 5: Replication, Maintenance, and Rearrangements of Genomic DNA
1/80
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
81 Terms
central dogma
DNA—→RNA—→Protein (polypeptide chain)
DNA polymerase
key enzyme in DNA replication, adds nucleotides in a 5’ to 3’ direction, creates the phosphodiester bonds between nucleotides
DNA polymerase gene family
Prokaryotes: I, II, III, IV, V
Eukaryotes: α, ε, δ
What is true of all DNA polymerases?
Synthesize 5’ to 3’
Can only extend pre-existing polynucleotide and must be bound to a template strand by complementary bases
What is the difference between DNA polymerase and RNA polymerase?
DNA polymerase requires a primer to initiate replication, RNA polymerase does not.
What are the differences between the leading and lagging strand?
Leading strand is continuous and only contains DNA primers. The lagging strand is discontinuous, broken up into Okazaki fragments, and contains RNA primers after ligation by DNA ligase.
What enzyme is responsible for removing the RNA primers from the ligated Okazaki fragments?
Exonuclease/endonuclease
Exonuclease
enzyme that cleaves RNA primers found at the ends of DNA templates (that were made up from Okazaki fragments).
Endonuclease
enzyme that cleaves RNA primers founds within the DNA template (that were made up of Okazaki fragments).
DNA polymerase I
Found in Prokaryotes. Fills in the gaps between Okazaki fragments, removing the RNA primers and replacing them with DNA.
DNA Polymerase III
Found in Prokaryotes. Synthesizing both the leading and lagging strands.
DNA Polymerase δ
Found in Eukaryotes. Fills in gaps between resulting from RNase H’s removal of RNA primers (synthesizes lagging strand).
RNase H
Found in Eukaryotes. An enzyme that degrades the RNA strand of RNA–DNA hybrids, and 5′ to 3′ exonucleases.
DNA Polymerase ε
Found in Eukaryotes. Synthesizes leading strand.
What do all cells need to initiate replication?
Origin of Replication (ori)
What enzyme do all Eukaryotic cells need for replication?
Primase (RNA primer)
DNA Polymerase α
Found in Eukaryotes. Initiates DNA replication by synthesizing short RNA–DNA primers that DNA polymerase δ or ε later extend.
Helicase
enzyme that catalyzes the unwinding of a DNA helix; breaks the hydrogen bonds between complementary base pairs (Requires ATP)
SSB (single-stranded DNA binding protein)
bind to the opening of replication fork to prevent unwound strands from bonding back together
sliding clamp protein (PCNA)
holds DNA polymerases in place it slides down the strands
clamp loading protein (RFC)
opens up PCNA and loads it onto/around the DNA strand (Requires ATP)
topoisomerase
relaxes supercoiled DNA by cutting it and ligating it back together without DNA ligase
In prokaryotes, what is the ori called and what does it do?
Initiator protein, it recruits enzymes necessary for replication (i.e. helicase, topoisomerase, etc.)
Eukaryotes have ______ ori because of the amounts of genes their DNA codes for.
multiple
Why does the leading strand synthesize faster than the lagging strand?
There is a higher occurrence of ori on the leading strand compared to the lagging strand.
In eukaryotes, what is ori called and what does it do?
Origin of Replication complex, made up of ARS and ACR, similarly recruits enzymes requires for replication.
In eukaryotes, how is DNA replication completed?
Uses telomerase to extend original DNA ends, using telomeres simple sequence repeats, then signaling standard replication machinery to fill in the complementary strand, so that the duplicated strand contains the desired sequence without shortening the chromosome; extends by repeating sequence prior to extension, makes a complimentary strand from telomerase’s RNA template, thus keeping the original DNA’s length in tact.
Telomerase
Enzyme responsible for extending DNA template
Is a specialized reverse transcriptase
Carries it’s own RNA template (complementary to telomeric sequence)
What happens to telomerase as you age?
Activity decreases causing cell death and senescence. Also indicates that telomeres are, in fact, shortening. (i.e. skin cells becoming senescent, ergo your skin gets thinner, less elastic, and wounds take longer to seal)
What happens if you have excessive telomerase activity?
Aids the rapid replication of cells, increasing the potential for cancerous tumors.
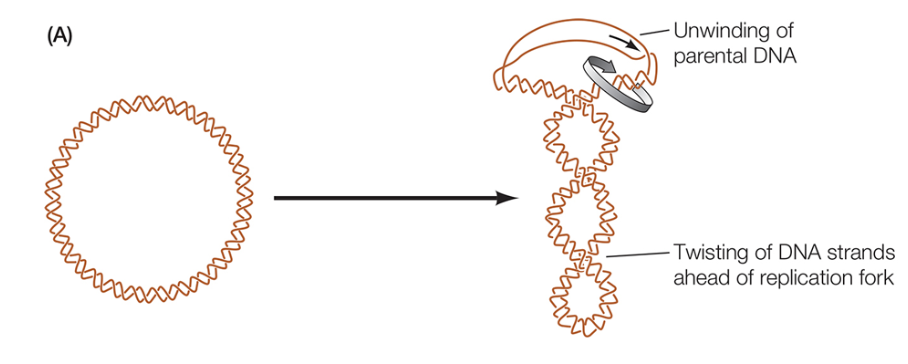
Which enzyme is pictured in this diagram?
Topoisomerase
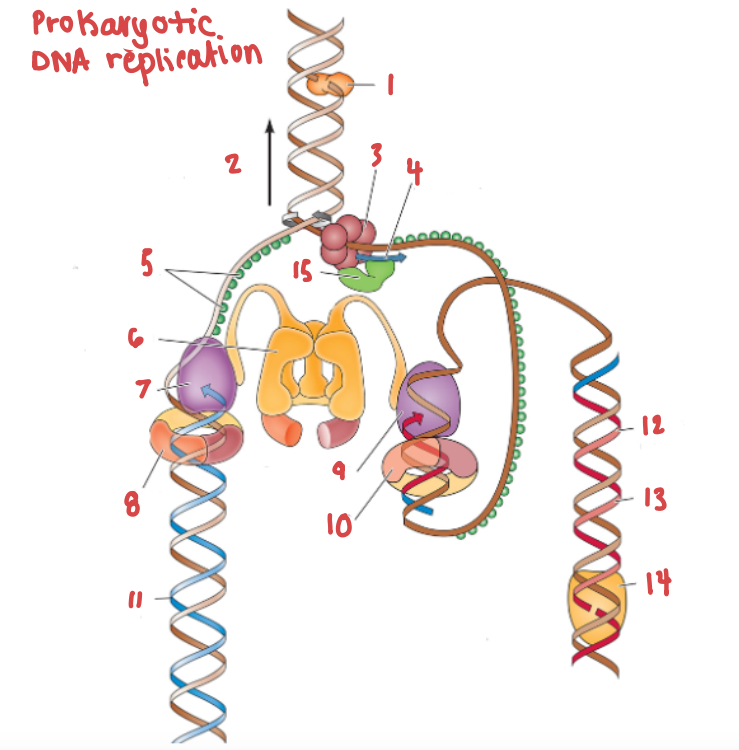
What is the name of the enzyme labelled 1?
Topoisomerase
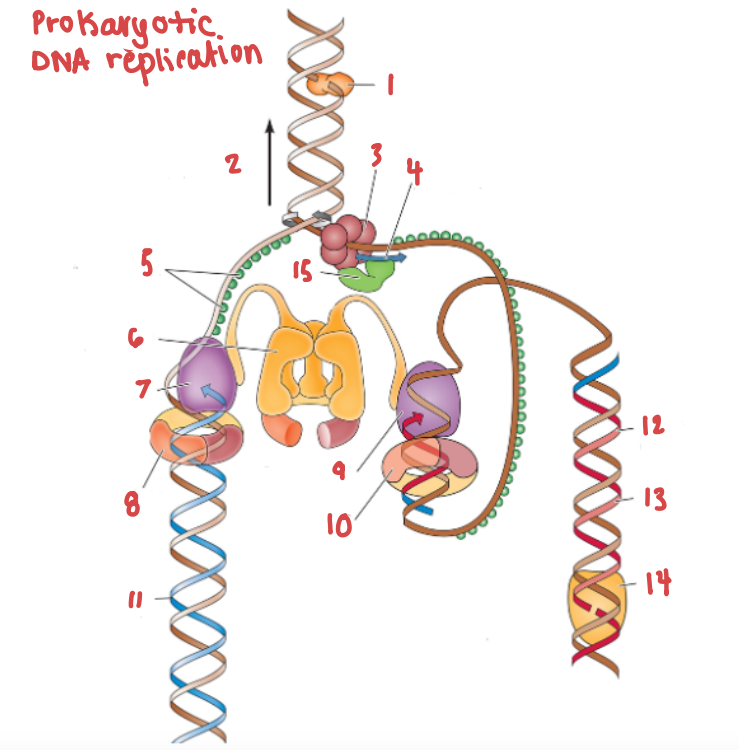
What is demonstrated by the arrow labelled 2?
Replication fork movement
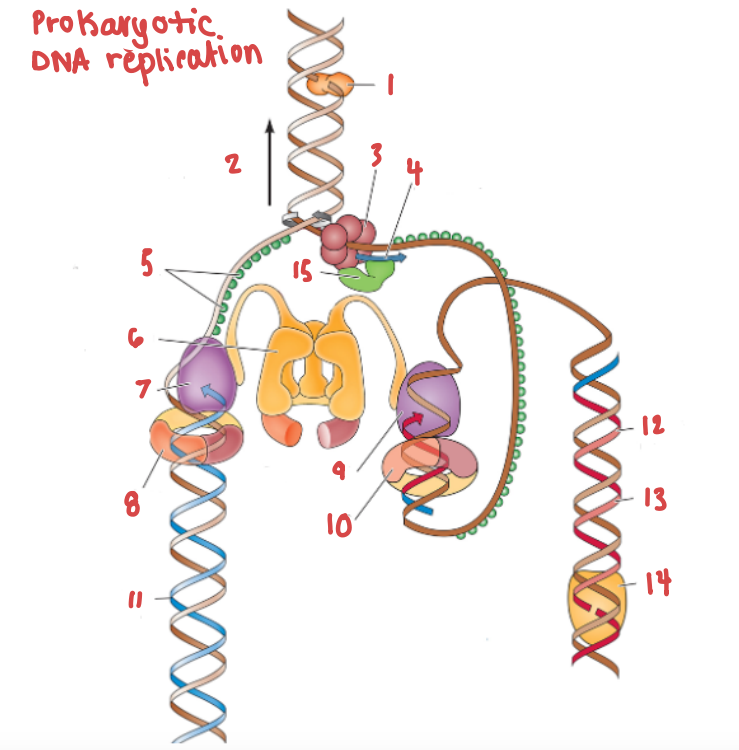
(3) is responsible for the unwinding of a DNA helix
helicase
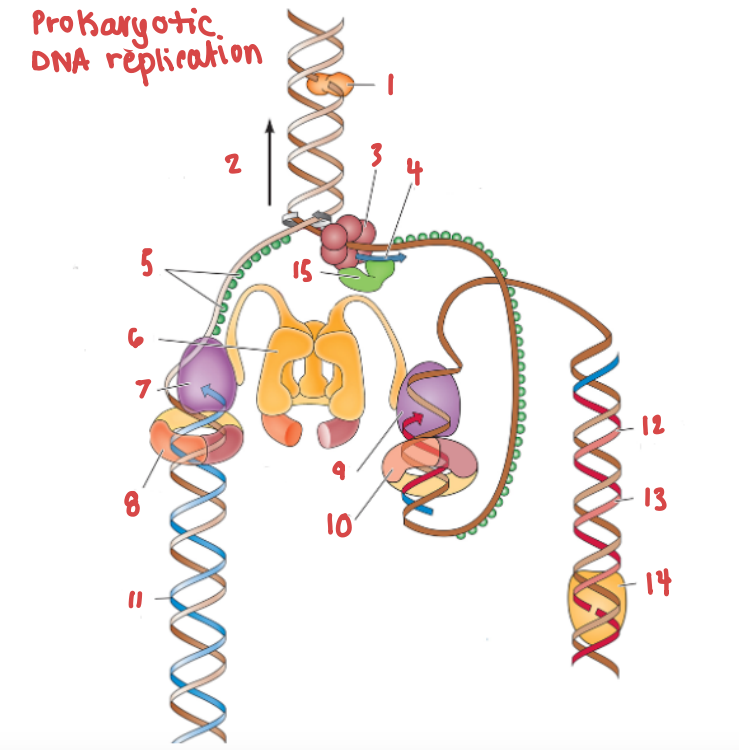
(15) and (4) work in tandem to create Okazaki fragments on the lagging strand.
Primase and RNA primer work in tandem to create Okazaki fragments on the lagging strand.
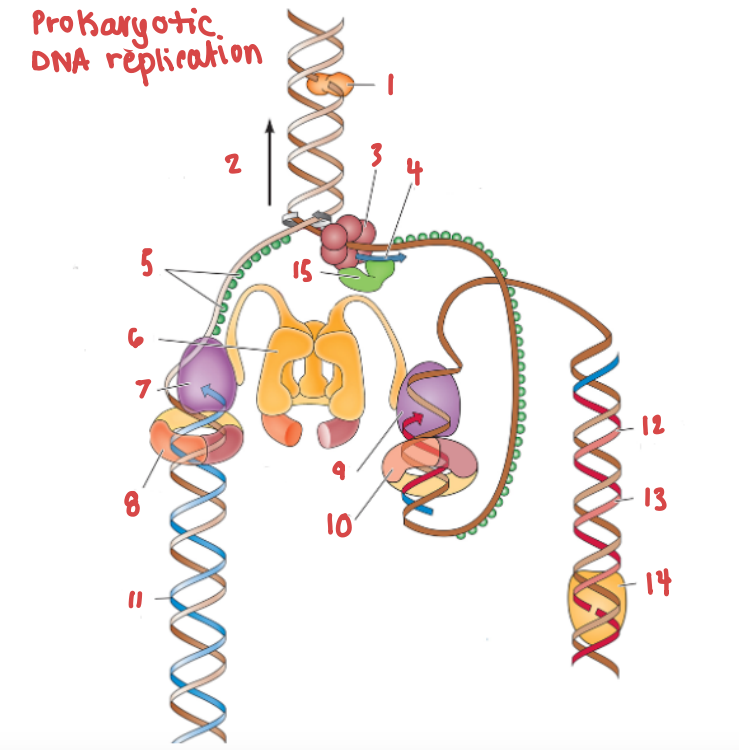
What is (5) and what is it doing?
SSB, preventing the strands from bonding together again.
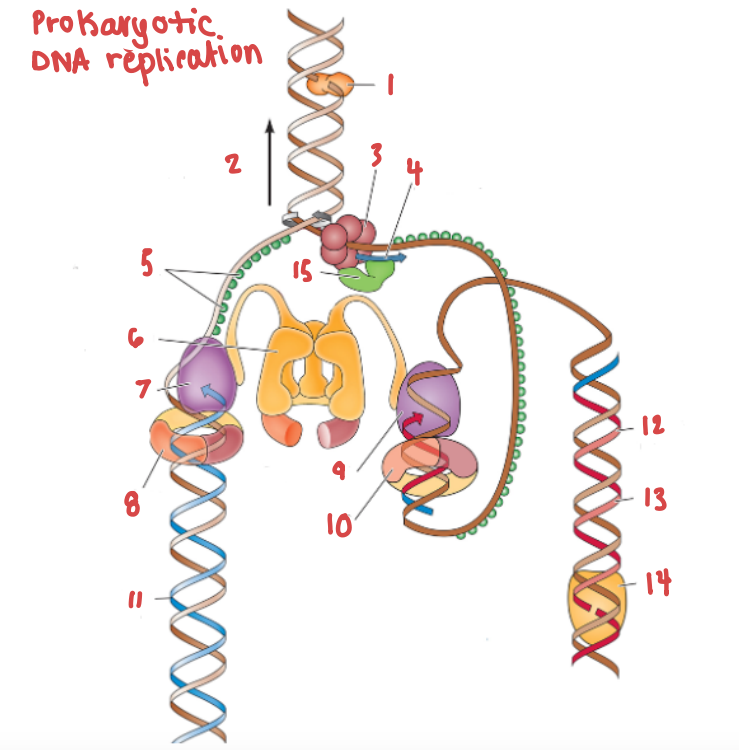
What is (6) and what is it doing?
Clamp loading protein, using ATP to open up PCNA and load it onto the strands.
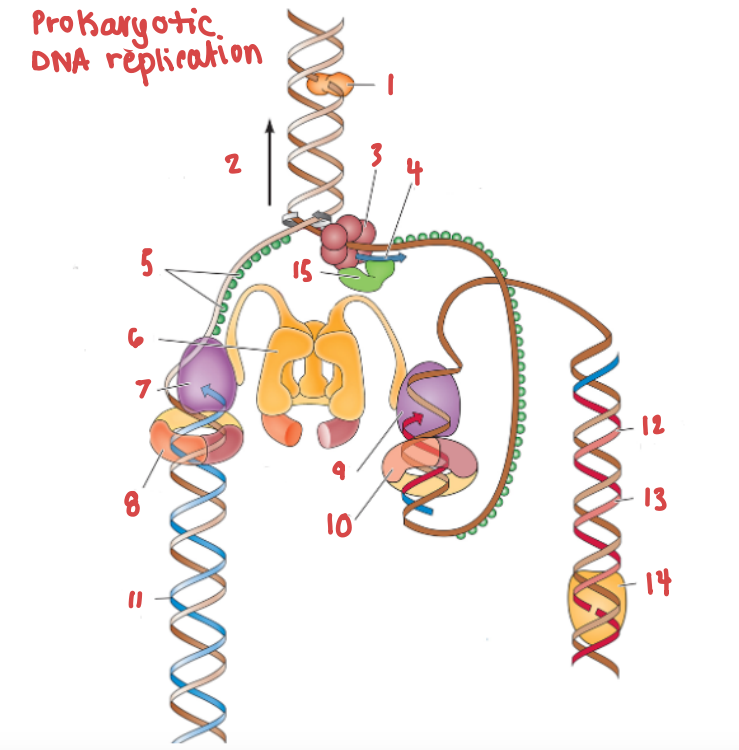
What is (7) and what gene family does it belong to?
DNA Polymerase III | DNA Polymerase
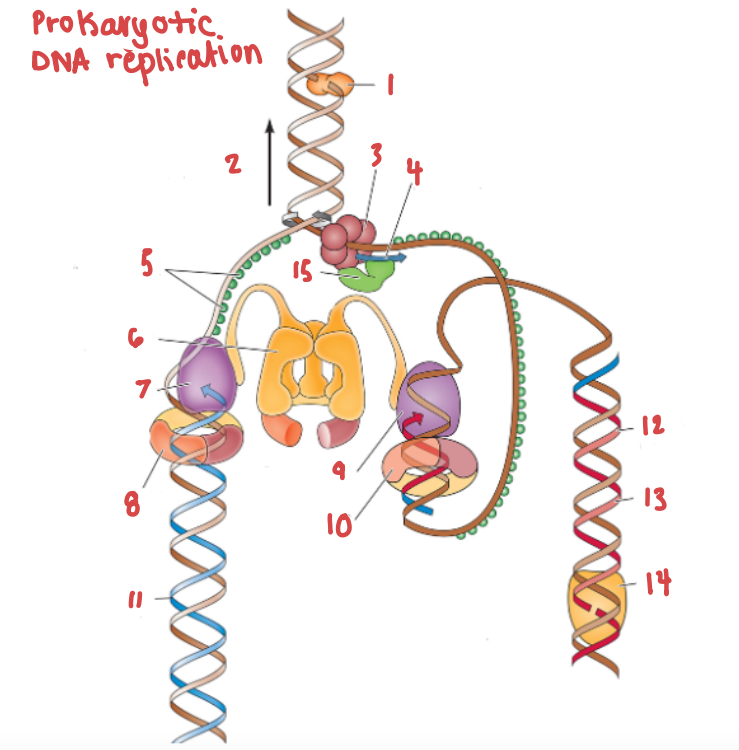
What are (8) and (9), and what are they doing to (7) and (10) respectively?
PCNA (sliding clamp proteins), and they are holding DNA polymerase III in place as it synthesizes the complementary strand.
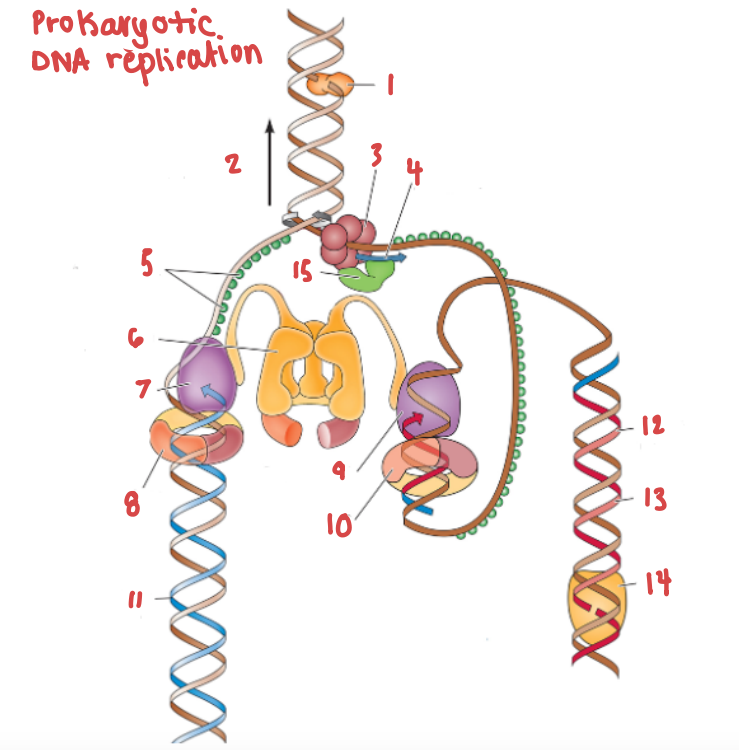
(11) is a continuous strand of complementary base pairs.
Leading strand
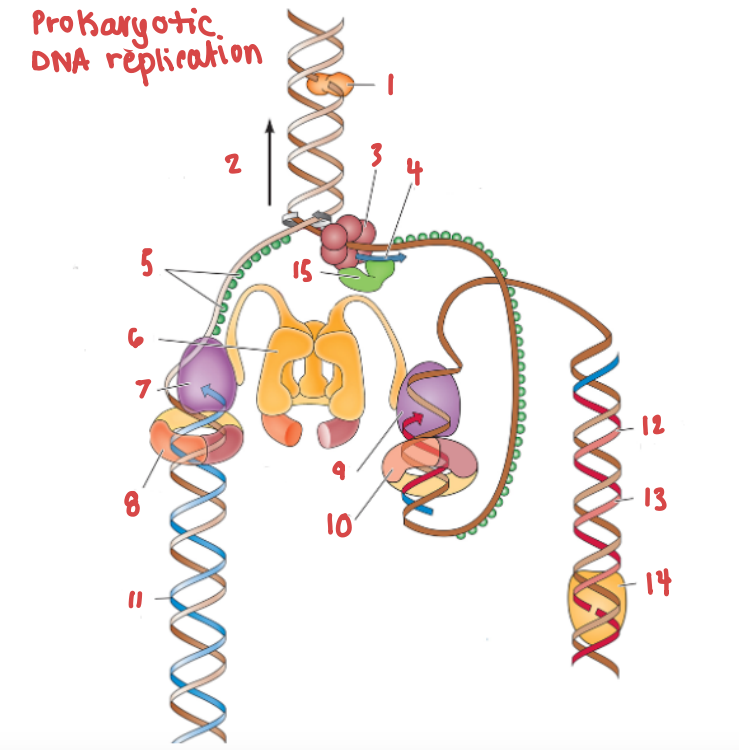
(12) make up (13), which are made into a continuous strand by (14).
Okazaki fragments make up the lagging strand, which are made into a continuous strand by DNA ligase.
In E. coli, the role of DNA polymerase I is to _______ while in mammalian cells this job is performed by _______.
degrade RNA primers; RNase H
You are interested in studying the role of RNA polymerase in DNA replication, so you treat cells with an inhibitor of RNA polymerase, after the initiation of replication, and determine its effects on DNA synthesis at the replication fork. How do you expect it to affect synthesis of the leading and lagging strands of DNA?
An inhibitor of RNA polymerase would block synthesis of the lagging strand by preventing synthesis of RNA primers. Synthesis of the leading strand would not be affected.
Etoposide is a small molecule that binds and inhibits topoisomerase II and has been used as a chemotherapeutic agent for cancer. Why would etoposide be effective against cancer?
Inhibiting topoisomerase II would be disruptive to DNA replication and the separation of intertwined DNA molecules during mitosis, which would be catastrophic to the proliferation of cancer cells.
What is the function of 3′ to 5′ exonuclease activity in DNA polymerases and what would be the consequence of mutating this activity of DNA polymerase III on the fidelity of replication of E. coli DNA?
3′ to 5′ exonuclease activity is required for the excision of mismatched bases during DNA replication so mutant E. coli lacking this activity would have a high frequency of mutations.
Why is the RNA molecule that exists in complex with telomerase essential to telomerase function in cells?
It associates with telomeric DNA via complementary base pairing and provides a template for the extension of telomeric repeat sequences by telomerase.
Which mechanism best ensures accurate DNA replication in prokaryotes?
Proofreading by DNA polymerase III
Which mechanism best ensures accurate DNA replication in eukaryotes?
Proofreading by DNA polymerase ε and δ
What does DNA polymerase detect during proofreading?
Mismatched deoxynucleotides (dNTPs). If it’s a purine-purine or pyrimidine-pyrimidine, the DNA polymerase hits “a bump in the road” due to the misshapen chemical structure.
DNA polymerase III, ε, and δ exhibit _________ when correcting a mismatched base pair.
3’ to 5’ exonuclease activity
The need for proofreading by DNA polymerase likely evolutionarily pressured…
DNA polymerase’s dependency on an annealed primer and 5’ to 3’ directionality of synthesis.
Where does the energy for phosphodiester bonds come from?
Hydrolysis of triphosphate bonds in deoxynucleotides (dNTPs).
Why is DNA synthesized 5’ to 3’?
Each incoming deoxynucleotide (dNTP) provides its own energy and proofreading by exonuclease does not halt synthesis.
Why is DNA not synthesized 3’ to 5’?
If DNA pol. removed a wrong nucleotide at 5’ end":
Only one phosphate remains, therefore there is not enough energy to continue synthesis.
Synthesis would stop if any errors are encounters.
AND
Evolutionary pressure favored 5’ to 3’ synthesis because it allowed proofreading and continuation of synthesis.
What can damage DNA?
UV Light
Cytosine deamination
Chemical adducts (i.e. chemical accumulation for smoking)
Radiation
How can UV light damage DNA?
Formation of pyrimidine dimers. Most commonly thymine-thymine (T-T) dimers, where adjacent thymine bases covalently bond to each other, forming a “knot” in the DNA strand.
How does cytosine deamination damage DNA?
Converts cytosine to uracil, creating a mismatch between uracil and guanine, leading to a point mutation. The removal of a amino group from cytosine is what converts it to uracil. Causes mutation amplification as DNA replicates because DNA polymerase will now read the uracil base and pair it with an adenine leading to a permanent point mutation.
How do chemical adducts damage DNA?
Bulky chemical groups covalently bond to the DNA helix, altering its structure and interfering with replication and transcription. Either lead to stalling synthesis, skipping lesions in the strand, or inserting the wrong bases. Leads to mutations and possible tumors if the bulky chemical is a carcinogen.
How does radiation damage DNA?
Causes double-strand breaks in the DNA which can result in mutations, cell death, and improper cell function. Capable of oxidizing DNA bases (dNTPS), breaking DNA strands, and creating cross-links between DNA strands or between DNA and other molecules in the cell.
What are some DNA repair mechanisms?
Proofreading
Mismatch Repair (MMR)
Base Excision Repair (BER)
Nucleotide Excision Repair (NER)
Translesion Synthesis (TLS)
Non-Homologous End Joining (NHEJ)
Which mechanism fixes a wrong nucleotide during replication?
Proofreading
Proofreading only happens when catalyzed by which enzyme(s)?
DNA polymerase and 5’ to 3’ exonuclease.
Proofreading…
removes mismatches immediately during synthesis.
Mismatch repair (MMR) fixes…
replication errors missed by proofreading.
Mismatch repair (MMR) requires which enzyme(s) for repair in bacteria?
MutS
MutL
MutH
The following enzymes are responsible for what part of MMR in bacteria? MutS, MutL, MutH.
MutS detects the mismatch pair or small insertion/deletion loops missed during DNA pol. proofreading.
MutL binds to MutS and coordinates by recruiting enzymes for the mismatch correction, also activating MutH.
MutH is activated by the MutS-MutL complex, then the endonuclease nicks or cuts the strand, providing an entry point for the enzymes responsible for correct.
Mismatch repair (MMR) requires which enzyme(s) for repair in eukaryotes?
MSH
MLH Homologs
The following enzymes are responsible for what part of MMR in eukaryotes? MSH and MLH Homologs.
MSH detects the mismatch pair or small insertion/deletion loops missed during DNA pol. proofreading.
MLH binds to MSH and coordinates by recruiting enzymes for mismatch correction, helping to identify the newly synthesized strand and initiating excision.
Base Excision Repair (BER) fixes…
Small lesions (C—→U, oxidized bases). Single-base damage caused by spontaneous or chemical modification.
Base Excision Repairs (BER) requires which enzyme(s)?
DNA glycosylase
AP endonuclease
DNA polymerase
DNA ligase
The following enzymes are responsible for what part of BER? DNA glycosylase, AP endonuclease, DNA polymerase, DNA ligase.
DNA glycosylase detects and removes the damaged or inappropriate base by cleaving the N-glycosidic bond, leaving behind an abasic (AP) site.
AP endonuclease recognizes the abasic site and cuts the DNA backbone next to it, creating an entry point.
DNA polymerase inserts the correct nucleotide(s) into the gap.
DNA ligase seals the nick in the sugar-phosphate backbone, restoring the DNA strand.
In Base Excision Repair (BER), which enzyme creates an AP site for precise, small-patch repair?
DNA glycosylase removes the damaged base, creating an AP site (apurinic/apyrimidinic).
Nucleotide Excision Repair (NER) repairs…
Bulky, helix-distorting DNA lesions (e.g., thymine dimers from UV light, chemical adducts).
Nucleotide Excision Repair (NER) requires which enzymes?
Endonuclease
DNA polymerase (ε or δ in eukaryotes & I in prokaryotes)
DNA ligase
The following enzymes are responsible for what part of NER? Endonuclease, DNA polymerase, DNA ligase.
Endonuclease recognizes bulky, helix-distorting lesions and cuts the DNA on both sides of the damage, excising the fragment.
DNA polymerase synthesizes new DNA to fill the resulting gap using the undamaged strand as a template.
DNA ligase seals the remaining nick in the sugar-phosphate backbone, fully restoring DNA integrity.
Translesion Synthesis (TLS) repairs…
DNA lesions that block normal replication (e.g., thymine dimers, abasic sites) by using specialized error-prone DNA polymerases to bypass the damage.
Translesion Synthesis (TLS) requires which enzymes?
Specialized translesion DNA polymerase.
Eukaryotes: Pol η, Pol ι, Pol κ, Pol ζ
Prokaryotes: Pol IV, Pol V
PCNA (ubiquitinated) – recruits TLS polymerases to the stalled fork