Genetics Exam 1: Lec. 3 (Human Genome Project)
1/77
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
78 Terms
Sanger developed techniques for sequencing DNA. What did he sequence?
Insulin
Which is the easiest to sequence: RNA, DNA, or proteins?
DNA- RNA is very unstable
Philip Sharp of MIT won a Nobel prize for the discovery that the sequence of mRNA differs from that of genomic DNA. Why is this?
non-coding regions (introns) have been removed that separate coding regions (exons)
what is used to convert mRNa into cDNA?
reverse transcriptase
Alternative splicing produces more than one mRNA. On average how many distinct proteins are made per human gene?
3
Carl Woese sequenced small subunit ribosomal RNAs and discovered a third kingdom of life called ___________.
Archaea
Archaea look like bacteria. What is the difference between the two?
Archaea live under extreme conditions such as high temperature.
Why was bacteria sequenced first?
their genomes are relatively small
The genome of Porphyromonas is a 2 Mbp circle. Why is Porphyromonas important for dental students?
Is one of the bacteria implicated in plaque formation and plays a role in periodontal progression
T/F Bacteria are haploid and so they obey Mendelian genetics.
False: Bacteria are haploid and so do not obey Mendelian genetics.
Bacteria exchange DNA by what three ways?
1) transformation
2) transduction
3) conjugation
Whole genome sequences of bacteria showed that there is massive ___________ __________ ____________ in the common ancestor to all cells and in bacteria now.
horizontal gene transfer
T/F Whole bacteria were made into mitochondria and chloroplasts.
True
horizontal gene transfers sets of genes that encode what two proteins in Salmonella?
1) secretory systems that allow Salmonella to secrete toxins into the host cell
2) proteins that confer antibiotic resistance
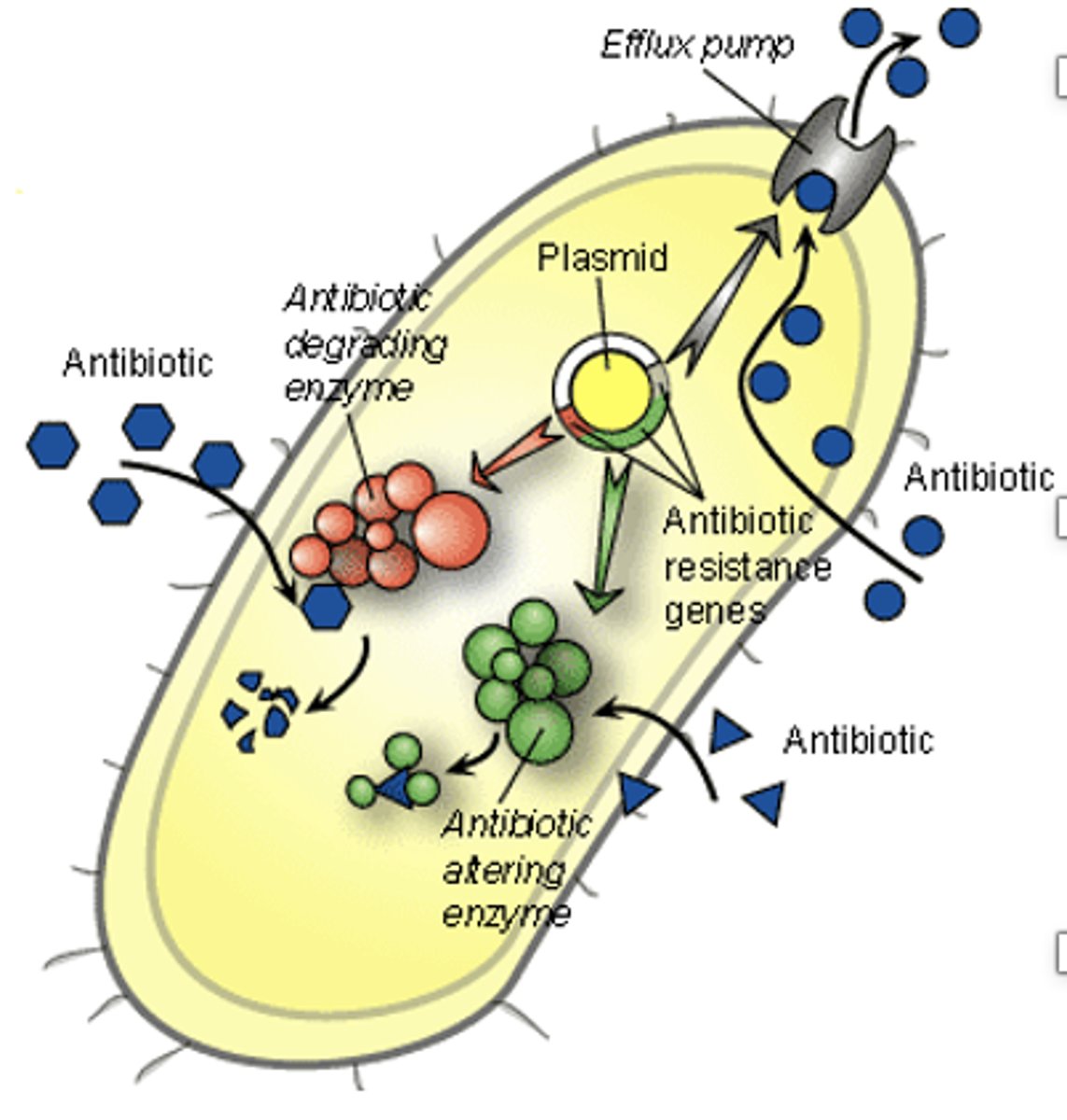
Microbial genome sequences make it possible to identify viruses and bacteria on our hands that may transmit infectious disease, as well as bacteria in our intestines (microbiome). What does the microbiome affect?
our general state of health and immune system
SARS-CoV-2 has an RNA genome, so it is diagnosed by ___-_____, not by PCR.
RT-PCR
SARS-CoV-2 makes a single Spike protein and a handful of enzymes. What are the two products targets for respectively?
1) single Spike proteins are target for vaccines and therapeutic antibodies
2) enzymes are targets for specific anti-viral drugs
Point mutations in the RNA genome make it possible to identify ___________, which are much more transmissible and so are targeted by new booster available now.
variants
the the following three steps necessary for preparing mRNA vaccines in order:
1 wrapping mRNA in a lipid coat to stabilize it and get it into the cell
2 nucleoside modifications that suppress the immunogenicity of RNA
3 stabilize the antigen expressed
2 > 1 > 3
2 nucleoside modifications that suppress the immunogenicity of RNA
1 wrapping mRNA in a lipid coat to stabilize it and get it into the cell
3 stabilize the antigen expressed
The RNA sequence of SARS-CoV-2 is used to diagnose COVID-19 infection (RT-RNA) and to design ___________ (Spike protein) or ____________ (e.g. Paxlovid that inhibits the protease).
vaccines (Spike protein)
anti-viral drugs (e.g. Paxlovid that inhibits the protease).
Studies of the human microbiome have led to efforts to prevent what thwo things?
1) spread of disease (hand sanitizers)
2) treatment of antibiotic-associated diarrhea (stool transplants).
•Microbial genome sequencing showed the importance of HGT for what three bacterial processes?
1) evolution
2) pathogenesis
3) drug resistance.
Where are the vast majority of mitochondrial proteins encoded and produced?
nucleus (encoded), cytosol and ribosomes (produced)
Mutations of proteins in the mitochondrial respiratory chain causes what?
hereditary optic neuropathy and vision loss
In addition to introns and exons, genes contain regulatory elements that control what?
mRNA expression
what percent of the human genome is repetitive or "junk" DNA
50%
repetitive or "junk" DNA includes what two repeats?
SINEs and LINEs
What is associated with euchromatin that is actively transcribed?
SINEs
What is associated with silent heterochromatin?
LINEs
Introns are much bigger than exons, so that less than _______% of the genome codes for proteins.
2%
Junk DNA is not entirely useless. What is derived from envelop proteins of retrovirus-like elements?
Syncytins
Syncytins are very abundant in the placenta and cause cells to fuse and form what?
syncytial trophoblast (outer placenta)
It appears that a retrovirus contributed to the development of placental mammals (Synctins). Why are non-viral examples of horizontal gene transfer (HGT) in mammals are rare?
the egg and sperm are protected from bacterial donors
Why proposed the following ideas:
1. Extensive sequencing of short DNA clones will eventually lead to large assemblies of overlapping sequences.
2. Proposed common disease/common variant hypothesis, which is the basis for genome-wide association studies (GWAS).
Eric Lander (probes dont need to know his name but good to know his "big ideas")
Whole exome sequencing (WES), which sequences exons that code proteins, is used to diagnose what?
genetic disorders causing birth defects and many other diseases
__________ _________ ___________ is cheaper and faster than whole genome sequencing and discovers rare or de novo mutations not detected by SNP arrays
Whole exome sequencing (WES)
Svante Pääbo, winner of the 2022 Nobel prize in Medicine, sequenced DNA from old human bones and showed that genomes of Europeans and Asians contain ~2% of what DNA?
Neanderthal DNA
____________, an extinct human species identified from a minute bone fragment (arrow), also contributed to our genome.
Denisovans
Where is DNA best preserved
Teeth
_______________ gave to modern humans viruses and genes encoding proteins to combat the viruses
Neanderthals
Genome sequences of thousands of individuals (ancient and contemporary) leads to what?
new maps of population flows that are more complicated than previously thought
T/F Genome sequences in public databases led to the arrest of the Golden State Serial Killer.
True
Sequencing of ancient genomes has confirmed human migration from where?
Africa
There are five methods to annotate a protein-encoding gene. Fillin' the following blank about one of the five methods:
Determine all possible _______ by deep sequencing of RNAs.
exons (protein products)
There are five methods to annotate a protein-encoding gene. Fillin' the following blanks about one of the five methods:
Determine ____________-____________ expression in humans or mice and determine the effect of __________-________ in mice or cultured cells (e.g., using CRISP/Cas9).
tissue-specific, knock-out
There are five methods to annotate a protein-encoding gene. Fillin' the following blanks about one of the five methods:
Express gene in bacteria or yeast to make ____________ __________ to test function and/or identify inhibitors.
recombinant protein
There are five methods to annotate a protein-encoding gene. Fillin' the following blank about one of the five methods:
Identify ____________ associated with cancer.
mutations
There are five methods to annotate a protein-encoding gene. Fillin' the following blank about one of the five methods:
Examine single nucleotide polymorphisms (SNPs) in human populations to see if there is an effect on susceptibility to _____________ ___________.
common diseases (hypertension, Alzheimer's, diabetes, etc.).
Jennifer Doudna won a Nobel prize in 2020 for adapting a bacterial system for fighting viruses into an extremely efficient system for engineering genomes of nucleated cells. Guide RNAs are engineered to bind to genomic DNA, which is cleaved by what?
Cas9 endonuclease
Once cleaved by Cas9 endonuclease both copies of a gene in a diploid organism may be __________ or genes can be ___________.
deleted (knockout)
modified (knockin)
CRISPR/Cas9 can be used to do what three things according to the lecture?
1) modify cultured human cells 2) fertilized eggs to produce to produce transgenic animals
3) and to modify tissues in adult mice
T/F there is a concern that CRISPR-Cas9 will be used to modify human germ line because it is so easy to use.
True
Henrietta Lacks (left) had cervical cancer from which the first immortalized human cell line (HeLa) was derived by George Gey at Johns Hopkins in 1951. Why were the cells immortal?
They had overreactive version of telomerase which prevented shortening of chromosome telomere and cellular aging
____________ cancer is caused by the human papilloma virus.
Cervical cancer
Whole genome sequencing of the HeLa cells shows HPV integrated in front of the _______ oncogene, which encodes a transcription factor that regulates growth.
MYC oncogene
T/F Sequences of tumor cells show mutations in cancer-associated proteins such as human p59 protein.
False: Sequences of tumor cells show mutations in cancer-associated proteins such as human p53 protein.
On average each cancer has how any important mutations in cancer-associated proteins.
3
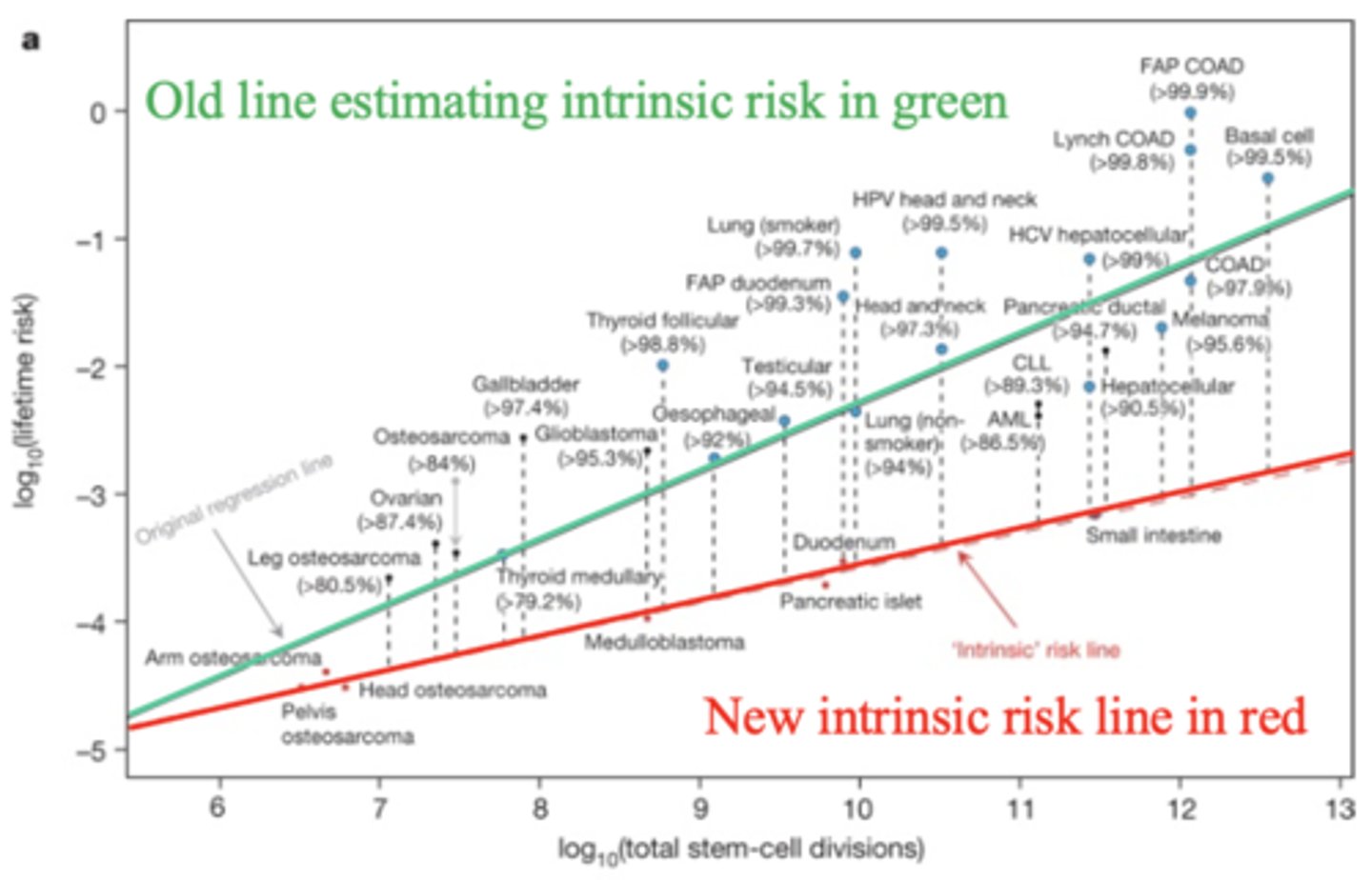
The lifetime risk of cancer increases with the total number of what? This explains why old persons have cancers in tissues that rapidly turnover.
stem cell divisions (exceptions are due to genetics of environmental factors)
Reinterpretation intrinsic risk suggests a much greater contribution for ____________ in cancer.
environment
Linkage studies that use Mendelian genetics are most effective in disorders, in which disease alleles are anticipated to have a ________ effect but occur ______________.
large, infrequently
Genome-wide association studies (GWAS), which use single nucleotide polymorphisms (SNPs), are most effective for the detection of alleles that occur ___________ but have a ___________ effect
frequently, small
De novo mutations, are discovered by whole genome sequencing (WGS) or whole exome sequencing (WES) and have ________ and ________ effects.
big and bad
Eric Lander's idea that common polymorphisms explain common diseases is not supported by data that most of the non-HLA genes associated with type 1 diabetes discovered by GWA studies of SNPs have ___________ effects.
small
GWAS to determine the cause of skin pigmentation in Africans (and everyone else) has been quite successful. 30% of the variation is explained in ___ genes, including one that causes mice to have grey rather than yellow hair.
6
Common diseases may be explained by rare variants discovered by whole genome sequencing. For example, 16% of individuals with severe mental retardation have a _____ _________ gene mutation that explains their condition.
de novo
Terminology: A theory that many common diseases are caused by common alleles that individually have little effect but in concert confer a high risk.
Common disease-common variant hypothesis
Terminology: A disorder in which the cause is considered to be a combination of genetic effects and environmental influences.
multifactorial (complex) disease
Terminology: A technique for sequencing a gene in several thousand subjects, typically with the use of high-throughput sequencing.
Deep resequencing
Terminology: A relationship that is defined by the nonrandom occurrence of a genetic marker with a trait, which suggests an association between the genetic marker (or a marker close to it) and disease pathogenesis.
Genetic association
Terminology: A relationship that is defined by the coinheritance of a genetic marker with disease in a family with multiple disease-affected members.
Genetic linkage
Define: A series of polymorphisms that are close together in the genome. The distribution of alleles at each polymorphic site is nonrandom: the base at one position predicts with some accuracy the base at the adjacent position. Persons sharing a ______ are related, often very distantly.
Haplotype
Terminology: A catalogue of common genetic variation in humans compiled by an international partnership of scientists and funding agencies. Its goal was to determine the identity and length of haplotypes across the genome in different human populations.
HapMap
Terminology: A coordinated international effort that led to the consensus sequence of the human genome.
Human genome project
Terminology: A disorder caused by a mutation in a single gene (also called a mendelian disease).
Monogenic disease
Terminology: Variation within the genome that results from deletion or duplication (both referred to as copy-number variation) or from inversion of genomic segments. Although common large variants (of more than one kilobase) exist, the majority of such variants are rare.
Structural genomic variation
Terminology: A description of all DNA that is transcribed into RNA (messenger RNA, transfer RNA, microRNA, and other RNA species). The prevalence of a specific RNA sequence in a particular tissue may be proportionate to the relevance of that RNA species in the tissue.
transcriptome
Terminology: A whole-genome resequencing of 1000 subjects from the original and extended HapMap populations, which was started in 2008, with funding from an international research consortium.
1000 Genomes project