BIOL 200 Lecture 6: Purification, Detection and Characterization of Proteins
1/13
Earn XP
Description and Tags
skeet 4
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
14 Terms
Purifying Proteins
Separates proteins based on different properties
We can obtain proteins from cells
Protein separation through chromatography
place soluble protein sample in a liquid phase inside column
Sample will move through solid phase, different proteins move at different speeds
Gel Filtration Chromatography
separating proteins based on their size
Beads in solid phase will have small pores that interact with sample.
Large proteins will not fit through pores and will fall straight to the bottom
smaller proteins stuck in pores are then washed out and collected
Ion exchange chromatography
separating proteins based on charge
1. Beads (solid medium) are given a charge
Proteins with same charge will fall through, while proteins with opposite charge will bind
Proteins are then washed with solution alike to them in charge. weaker bonded proteins fall sooner because they are outcompeted for the bonds more easily.
Antibody affinity chromatography
Purify proteins based on antibody interactions
Any proteins that don’t bind to the antibodies on the beads fall through
Proteins that did bind (had correct antibodies) are then removed by lowering the pH to weaken the binding.
Electrophoresis
protein mixtures loaded in wells
voltage applied so proteins migrate from negative to positive side
Smaller proteins move through the pores faster
Direction of migration is determined by net charge (attracted or repelled to +)
Speed of migration determined by mass/charge ratio
What does SDS do in gel electrophoresis?
denatures proteins
hydrophobic tail of SDS interacts with hydrophobic amino acids inside protein.
gives all proteins uniform negative charge.This allows proteins to be separated based solely on their size during electrophoresis.
What is SDS - PAGE?
A type of electrophoresis used to study protein that infects phages
All proteins look the same, just different lengths
Immunoblot
Used to detect specific proteins in an SDS - PAGE sample
sample transferred to membrane that exposes protein surface
primary antibody detects an exposed protein
tagged secondary antbody binds to primary antibody (fluorescent or radioactive)
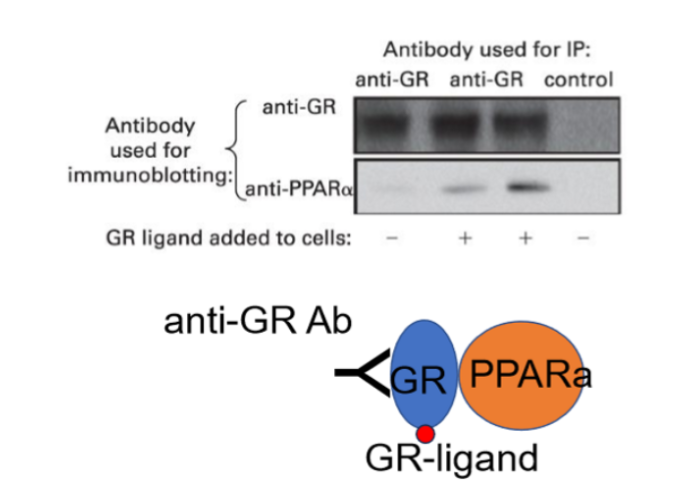
Co-Immunoprecipitation
Antibodies that only recognize certain proteins
used to purify other proteins in a complex that interact with it
Samples used are not treated, maintaining tertiary and quarternary structures
Immunoprecipitate contains protein which carries a structure that antibodies recognize (epitope) and partner proteins
Antibody recognizes GR which interacts with a second ligand and PPARa
Immunofluorescence
antibodies used to locate proteins in cells and tissues
Cell put in solvent that ensures structure preservation
Primary antibodies detect proteins
Tagged secondary antibodies attach to primary antibodies, allowing visualization under a fluorescence microscope.
Fluorescence
Electrons named fluorophores absorb photons, causing electrons to get excited and move to a higher orbital
when they return to ground state they emit light
excitation-emission cycles are oxidized to stop producing fluorescence.
How does a fluorescent microscope work?
Uses a filter to block and direct light
Light hits filter and reaches dichroic mirror that reflects green light and lets red light through
We view green fluorescence through objective lens under the microscope.
Red light is passed through the emission filter.
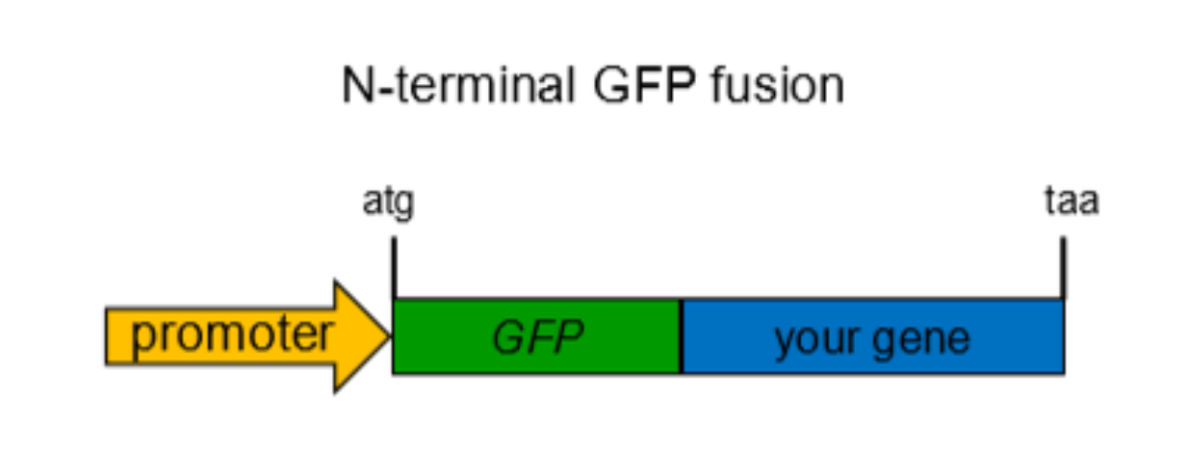
Gene fusion
done by cloning, it is used to mark proteins of interest
Endogeneous fusion involves homologous recombination. C or N terminal is chosen depending on which end the protein is.
All resulting proteins will be marked by fluorescent protein.
Ex. GFP keeps cells alive to allow for real time tracking of proteins we are interested in.Gene fusion is a technique where two genes are joined together, often used to attach a reporter gene, like GFP, to a target protein, enabling visualization and tracking within living cells.