Lecture 13 - Nucleotides and nucleic acids
1/57
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
58 Terms
what are nucleic acids?
Nucleic acids are polymers of nucleotides and taken together
they fulfill a diverse group of roles in the cell:
what are five functions of nucleotides?
Precursors of RNA and DNA
High energy sources (ATP, GTP)
Regulatory signals (cAMP)
Coenzymes (FAD, NAD, NADP)
High energy intermediates in metabolism
eg. UDPglucose (CHO), CDP-acylglycerol (lipid)
what are the two types of nucleic acids?
Dna - deoxyribonucleic acid
RNA - Ribonucleic acid
characteristics of dna (3)
Sugar: Contains 2'-deoxy-D-ribose (notice the missing OH on the 2' carbon).
Function: Stores genetic information (it’s the cell’s long-term information archive).
Key feature in structure: Double-stranded helix.
characteristics of RNA
Sugar: Contains D-ribose (has OH on the 2' carbon).
Functions:
Carries genetic information (like mRNA).
Has catalytic roles (e.g., ribozymes).
Plays regulatory roles (like RNA interference, miRNA, etc.).
Structure: Usually single-stranded.
✅ Tip: That OH group on the 2' carbon makes RNA less stable than DNA and more chemically reactive.
what is a nucleoside?
Nucleoside = Base + Sugar
Contains:
A sugar (either ribose or deoxyribose)
A nitrogenous base (purine or pyrimidine)
🔄 Think of it as a half-built nucleotide – it's missing the phosphate group.
what is a nucleotide?
Nucleotide = Nucleoside + Phosphate
Contains:
Sugar
Base
Phosphate group(s) (can be 1, 2, or 3 – like AMP, ADP, or ATP)
Nucleotides are the monomers that get linked into RNA and DNA.
what is prime notation?
The prime (′) notation is used for the sugar portion of nucleotides:
In a nucleotide, you have two parts that both contain carbon atoms:
A nitrogenous base (like adenine, cytosine, etc.)
A sugar (ribose or deoxyribose)
Both parts have their own carbon numbering.
To avoid confusion:
The carbons in the nitrogenous base are labeled without primes (e.g. C1, C2, etc.).
The carbons in the sugar are labeled with primes (e.g. C1′, C2′, C3′, C4′, C5′).
Nucleotides have nitrogenous bases that are either:
Pyrimidines (1 ring)
Purines (2 rings)
what are Pyrimidines (1 ring)
6-membered single-ring structure
Atoms are numbered 1–6
Examples: Cytosine, Thymine (DNA), Uracil (RNA)
what are Purines (2 rings) (3)
Fused 6-membered + 5-membered ring (total 9 atoms)
Atoms are numbered 1–9
Examples: Adenine, Guanine
what are the common purine bases?
Adenine (A)
Guanine (G)
➡ Found in both DNA and RNA
what are the common Pyrimidine Bases
Cytosine (C) – both DNA and RNA
Thymine (T) – only in DNA
Uracil (U) – only in RNA (replaces thymine)
what are Ribonucleotides?
RNA Building Blocks
Ribonucleotides = ribose sugar + base + phosphate
Sugar = D-ribose → has an –OH at the 2′ carbon
Linked via a β-N-glycosidic bond:
Purines (A, G): bond to N9
Pyrimidines (C, U): bond to N1
B-N glycosidic bond = The sugar (ribose or deoxyribose) uses its anomeric carbon (C1').
It forms a β-glycosidic bond with the nitrogen in a nitrogenous base (A, G, C, T, or U).
what are Deoxyribonucleotides?
DNA Building Blocks
Deoxyribonucleotides = deoxyribose sugar + base + phosphate
Sugar = 2′-deoxy-D-ribose (no –OH at the 2′ position — just H)
Also connected via β-N-glycosidic bond (same as RNA)
DNA nucleotides are Adenine, guanine, thymine and cytosine
Naming Patterns for nucleotides
Base | Nucleoside (RNA) | Nucleotide (RNA) | Nucleic Acid |
|---|---|---|---|
Adenine | Adenosine | Adenylate | RNA |
Guanine | Guanosine | Guanylate | RNA |
Cytosine | Cytidine | Cytidylate | RNA |
Uracil | Uridine | Uridylate | RNA |
Thymine | Thymidine or deoxythymidine | Thymidylate or deoxythymidylate | DNA (usually) |
is it true that nucleotides dont always have a single phosphate?
Nucleotides don’t always just have a single phosphate at the 5' position — they can be modified for signaling purposes.
what are the different structural forms of AMP (adenosine monophosphate) based on where the phosphate group is attached (4)
Molecule | Description |
|---|---|
Adenosine 5′-monophosphate (AMP) | Regular AMP, phosphate on 5′ carbon of ribose |
Adenosine 3′-monophosphate | Phosphate is attached to 3′ carbon |
Adenosine 2′-monophosphate | Rare, phosphate on 2′ carbon |
Adenosine 3′,5′-cyclic monophosphate (cAMP) | A cyclic nucleotide, phosphate forms a ring by linking both 3′ and 5′ hydroxyls of ribose |
What is cAMP?
cAMP stands for cyclic adenosine monophosphate — a cyclic nucleotide derived from ATP.
Function of cAMP: 🔔 It acts as a second messenger in signal transduction:
A first messenger like a hormone (e.g., epinephrine) binds to a receptor on the cell surface.
This activates adenylyl cyclase, which converts ATP → cAMP.
cAMP then:
Activates protein kinase A (PKA)
Triggers phosphorylation cascades
Regulates enzymes, ion channels, gene expression, etc.
what is a energy source for nucleotides?
Phosphate Groups = Energy Units
Nucleotides can have:
1 phosphate → monophosphate (e.g., AMP)
2 phosphates → diphosphate (e.g., ADP)
3 phosphates → triphosphate (e.g., ATP)
These phosphates are linked by high-energy bonds, especially the terminal phosphate bonds. Breaking these bonds releases energy used by the cell.
How is ATP used as a energy source
ATP (Adenosine Triphosphate)
The most common cellular energy carrier
Consists of:
Adenine base
Ribose sugar
3 phosphate groups (α, β, γ)
The energy comes from hydrolyzing the terminal phosphate bond, forming ADP + Pi or AMP + PPi
the energy released by breaking the high-energy phosphate bonds in nucleotide triphosphates is used to form phosphodiester bonds that link nucleotides together in DNA and RNA synthesis.
what are other energy molecules similar to atp? (3)
GTP – used in protein synthesis and signaling (Nbase = guanine)
CTP – used in lipid synthesis (Nbase = Cytosine)
UTP – used in carbohydrate metabolism (e.g., UDP-glucose) (Nbase = uracil)
GTP, CTP, and UTP are all nucleotide triphosphates, just like ATP — and each plays important roles in energy transfer, signaling, and nucleic acid synthesis.
how do nucleotides link to form DNA/RNA
Nucleotides link to form DNA/RNA through:
A phosphodiester bond is a covalent bond between:
The phosphate group on the 5′ carbon of one nucleotide, and
The hydroxyl (–OH) on the 3′ carbon of the next nucleotide's sugar.
Creates a sugar-phosphate backbone.
Think of it like beads on a string — the sugars are the beads and the phosphates are the strings between them.
what is the direction of strands?
5′ to 3′ Direction
Each strand of DNA or RNA has a 5′ end (with a phosphate) and a 3′ end (with a hydroxyl).
Sequences are always written 5′ → 3′.
DNA vs RNA structure
Feature | DNA | RNA |
|---|---|---|
Bases | A, T, G, C | A, U, G, C |
Sugar | Deoxyribose | Ribose |
Structure | Usually double-stranded | Usually single-stranded |
what are nucleotide units held together by?
Phosphodiester Bonding
Nucleotides are linked by phosphodiester bonds, which connect:
The 3′-OH of one nucleotide’s sugar
To the 5′-phosphate of the next
This creates a directional strand with:
A 5′ end (starting with a phosphate group)
A 3′ end (ending in a free OH)
How Sequences Are Written?
Always written from 5′ → 3′ (left to right)
For example:
pApCpGpTpA= ACGTA
“p” means phosphate; this notation just emphasizes the individual units.
what was known about dna structure in 1953?
DNA = genetic material (not protein!)
Chargaff's rules:
Amount of A = T
Amount of C = G
Shows base pairing symmetry
Watson & Crick proposed a 3D structure of B-DNA (the form believed to exist in chromosomes) in 1953 using this information and X ray diffraction data
what was watsons and crick 3d structure of dna?

what are dna strands help together by?
DNA strands are held together by hydrogen bonds between nitrogenous bases:
Pair | # of H-Bonds | Stability |
|---|---|---|
A–T | 2 | Less stable |
G–C | 3 | More stable (stronger pairing) |
These base-pairing rules explain Chargaff's observations:
%A = %T
%G = %C
what does it mean by Complementary Strands?
A sequence on one strand determines the sequence of the opposite strand.
For example:
5′-A T G C-3′
3′-T A C G-5′
This makes the strands different but complementary—a vital concept for replication and transcription.
whats the point of the Major and Minor Grooves
The twisting of the helix creates grooves:
Major groove: wider, more accessible → often targeted by DNA-binding proteins
Minor groove: narrower
These grooves run parallel to the backbone and allow proteins to “read” the sequence without breaking the helix.
what the dna structures dimensions
Base pair spacing:
0.34 nm (3.4 Å) between adjacent base pairs
1 full turn of the helix:
3.6 nm (36 Å) per turn
10.5 base pairs per turn
What is Base Stacking?
Bases in DNA are flat, planar, and hydrophobic, so they stack on top of each other within the helix.
what is this stacking stabilized by?
This stacking is stabilized by:
van der Waals forces
dipole–dipole interactions
This contributes to DNA helix stability, along with hydrogen bonding between base pairs.
how does the Hydrophobic Effect come into this?
Since bases are hydrophobic, stacking hides them from water, minimizing their contact with the aqueous environment (i.e., hydrophobic shielding).
This is thermodynamically favorable.
therefore, Base stacking is as important—if not more—than H-bonding in stabilizing the DNA double helix.
how are grooves made?
Because of the asymmetry in glycosidic bonds (base-to-sugar connections), the two backbones don't align evenly, creating:
A major groove (wider)
A minor groove (narrower)
function of grooves? (3)
Proteins (e.g., transcription factors) can bind to the major groove (more accessible) and read DNA sequences without unzipping the double helix.
This enables gene regulation and sequence-specific binding (like promoter recognition, enhancer binding, etc.)
Binding happens via H-bonding, van der Waals, and shape complementarity.
how many forms of DNA are there?
Three forms
A
B
Z
B-form DNA is the most common DNA under physiological conditions
Why DNA/RNA can twist and fold?
The sugar-phosphate backbone has 7 rotatable bonds (shown as red dots).
These allow for:
Twisting and bending
Folding into secondary (e.g., stem-loops) and tertiary (e.g., tRNA structure) conformations
contrast that with proteins, which have only 2 main rotatable bonds per residue (ϕ and ψ).
why does this twisting and folding matter?
These torsion angles enable supercoiling, helix formation, and complex folding of RNA (especially important for ribozymes or tRNA).
Also affects binding with proteins, enzymes, etc.
Twisting and folding are essential for nucleic acids to be stable, compact, and — in the case of RNA — functional. Without proper 3D structure, DNA couldn’t be stored or replicated correctly, and RNA couldn’t carry out its many roles in the cell.

What is DNA denaturation?
DNA isn’t static—it breathes, meaning small regions can unwind and rewind dynamically.
When temperature increases, thermal motion breaks the hydrogen bonds between base pairs, eventually causing complete strand separation.
What can increase Temp cause?
Denaturation = Loss of Base Pairing & Stacking
Base pairing and base stacking both stabilize DNA.
Melting disrupts:
H-bonds between base pairs
Stacking interactions between adjacent bases
This is called “melting”, even though it’s not melting in the usual physical sense.
What else causes denaturation?
pH extremes (very high or low) can also break hydrogen bonds
Enzymes like helicases separate DNA during replication
What happens when DNA melts?
As DNA strands separate:
Base stacking is lost
UV absorbance increases (more light is absorbed)
This increase in absorbance is called the hyperchromic shift
how can DNA melting be measured?
DNA melting can be measured using changes in absorption of UV light
Mononucleotides absorb UV light at 260 nm due to their aromatic rings
In double-stranded DNA, base stacking decreases absorbance → this is called the hypochromic effect
what is the hypochromic effect?
The hypochromic effect refers to the decrease in UV light absorbance (usually at 260 nm) that occurs when single-stranded nucleic acids become double-stranded (as in DNA helix formation).
why does the hypochromic effect occur?
It all comes down to base stacking and hydrogen bonding:
In single-stranded DNA or RNA:
The nitrogenous bases are unstacked and exposed.
They freely absorb UV light around 260 nm.
Result: Higher absorbance (hyperchromic state).
In double-stranded DNA (dsDNA):
The bases are hydrogen-bonded and tightly stacked inside the helix.
This restricts their ability to absorb UV light.
Result: Lower absorbance (hypochromic effect).
so what happens to uv absorbance when DNA/RNA denatures?
Heat breaks the hydrogen bonds and unravels the helix.
Bases become exposed → absorb more UV → absorbance increases.
This increase in absorbance during melting is called the hyperchromic shift.
The hypochromic effect occurs in both DNA and RNA,
whenever bases are stacked and hydrogen-bonded, reducing their ability to absorb UV light.
what is Melting Temperature (Tm)?
Tm = temperature at which 50% of the base pairs in DNA are denatured (single stranded) and half are still double stranded
It reflects thermal stability of the DNA
Higher GC content = higher Tm, because:
G≡C has 3 hydrogen bonds (vs. A=T with 2)
GC pairs also have stronger stacking
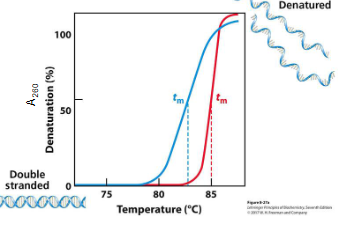
explain this graph
UV Absorbance & Tm:
A260 (absorbance at 260 nm) is plotted against temperature
As DNA denatures:
Absorbance increases (hyperchromic effect)
The sigmoidal curve reflects the cooperative nature of melting
The midpoint of this curve = Tm
why does more GC = Higher tm?
GC base pairs form 3 hydrogen bonds, while AT forms only 2 → more heat is needed to break GC pairs.
GC pairs stack better, contributing more to the helix’s stability than AT.
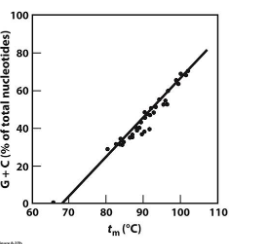
what does this straight line mean?
Tm increases linearly with %GC content.
This means base composition directly affects DNA thermal stability.
High GC DNA = More stable = Higher Tm
Final Summary: of tm
Tm = temperature at which half the DNA is melted
Measured by UV absorbance at 260 nm
More GC content = higher Tm due to:
More H-bonds
Stronger stacking
what happens during dna replication?
What Happens During Replication?
DNA duplicates before cell division
The two strands separate (unzipped)
Each serves as a template to synthesize a new complementary strand
A pairs with T, G pairs with C
Result = 2 identical DNA molecules, each with one parent and one new strand (called semi-conservative replication)
what is the Role of DNA Polymerases:
DNA polymerases are enzymes that:
Read the template strand
Add complementary nucleotides to form the new strand
what helps dna polymerase?
Assisted by helper proteins like:
Helicase (unzips DNA)
Primase (lays RNA primer)
Ligase (seals fragments)
why does accuracy matter?
Mistakes = mutations
High-fidelity polymerases and proofreading functions ensure low error rates