Cytoplasmic Membranes #2
5.0(2)
5.0(2)
Card Sorting
1/49
Earn XP
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
50 Terms
1
New cards
What are coated vesicles?
These are vesicles that bud from a membrane compartment that typically has a multi-subunit protein coat which promotes the budding process and binds specific membrane proteins
2
New cards
What are the 3 best known coated vesicles?
1. COP1
2. COP2
3. Clathrin-coated vesicles
3
New cards
COP1 function
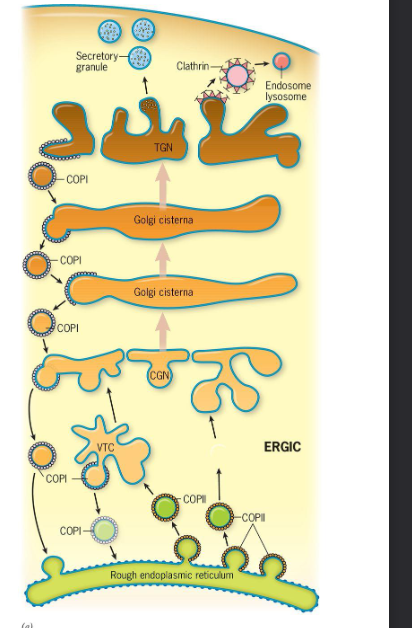
Move materials in a ^^retrograde^^ direction from the ERGIC and Golgi stack backward toward the ER and from ==trans Golgi cisternae== “backward” ==to cis Golgi cisternae==
4
New cards
COP 2 function
* Move materials ==from the ER== “%%forward%%” ==to the ERGIC== that the ERGIC is the intermediate compartment which is situated between the ER and Golgi Complex (COP=coat proteins)
* Goes to ER, then to golgi
* Goes to ER, then to golgi
5
New cards
Clathrin-coated vesicles
__Move materials from the Golgi to the TGN to endosomes, lysosomes, and plant vacuoles.__ They also __move materials from the plasma membrane to cytoplasmic compartments__ along the endocytic pathway and have also been implicated in trafficking from endosomes and lysosomes.
6
New cards
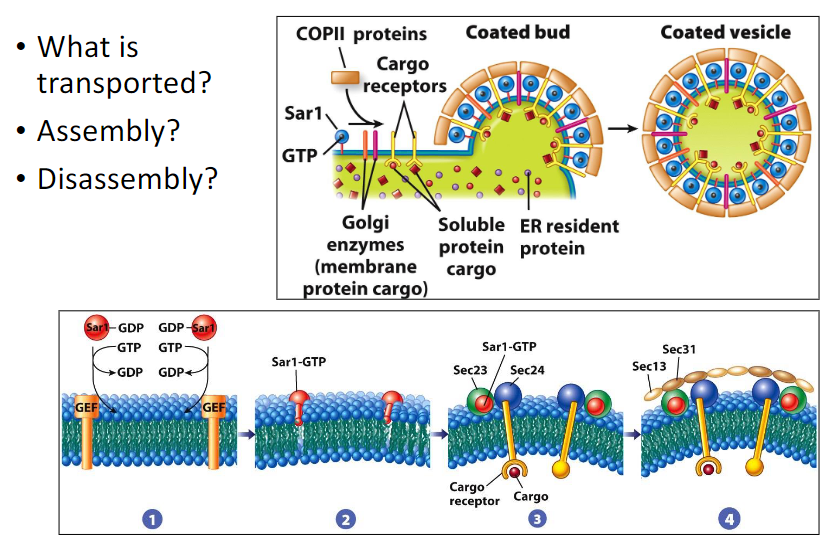
What types of proteins are transported with COP2 vesicles? How does the COP2 proteins (Sec 13, 23, 24, 31), Sar, GTP, GEF and cargo receptors interact during the formation of COP2 vesicles?
* Enzymes that act at later stages in the biosynthetic pathway (glycosyltransferase of the Golgi complex),
* Membrane proteins involved in the docking and fusion of the vesicle with the target compartment
* Membrane proteins that can bind soluble cargo (like secretory proteins)
* GTP, Sar, and GEF undergo a conformational change which causes its N-terminal alpha helix to insert itself into the cytosolic leaflet of the ER bilayer
* Membrane proteins involved in the docking and fusion of the vesicle with the target compartment
* Membrane proteins that can bind soluble cargo (like secretory proteins)
* GTP, Sar, and GEF undergo a conformational change which causes its N-terminal alpha helix to insert itself into the cytosolic leaflet of the ER bilayer
7
New cards
How are COP2 vesicles assembled and disassembled?
Find answer in book
8
New cards
G-Proteins
* GTP binding protein
* Molecular switches and timers
* Cell has a timing mechanism within it
* GTPase promotes binding to GTP
* GTP-binding proteins or G proteins are transmitting signals outside the cell which cause changes within the cell. They act as molecular switches which are %%on when binding GTP%% and ^^off when binding GDP^^.
* Molecular switches and timers
* Cell has a timing mechanism within it
* GTPase promotes binding to GTP
* GTP-binding proteins or G proteins are transmitting signals outside the cell which cause changes within the cell. They act as molecular switches which are %%on when binding GTP%% and ^^off when binding GDP^^.
9
New cards
Discuss, in detail, how COPI vesicles are able to help maintain the appropriate distribution of proteins in the Golgi and RER: retention
Retention: resident molecules that are excluded from transport vesicles are based mostly on the physical properties of the protein
10
New cards
Discuss, in detail, how COPI vesicles are able to help maintain the appropriate distribution of proteins in the Golgi and RER: retrieval
Retrieval: Escaped molecules back to the compartment in which they typically reside
11
New cards
Exactly how are these vesicles able to recognize the proteins they are supposed to transport?
* How do these maintain \\n proper distribution of \\n RER and Golgi proteins?
* Look in book
* @@How does recognition occur?@@
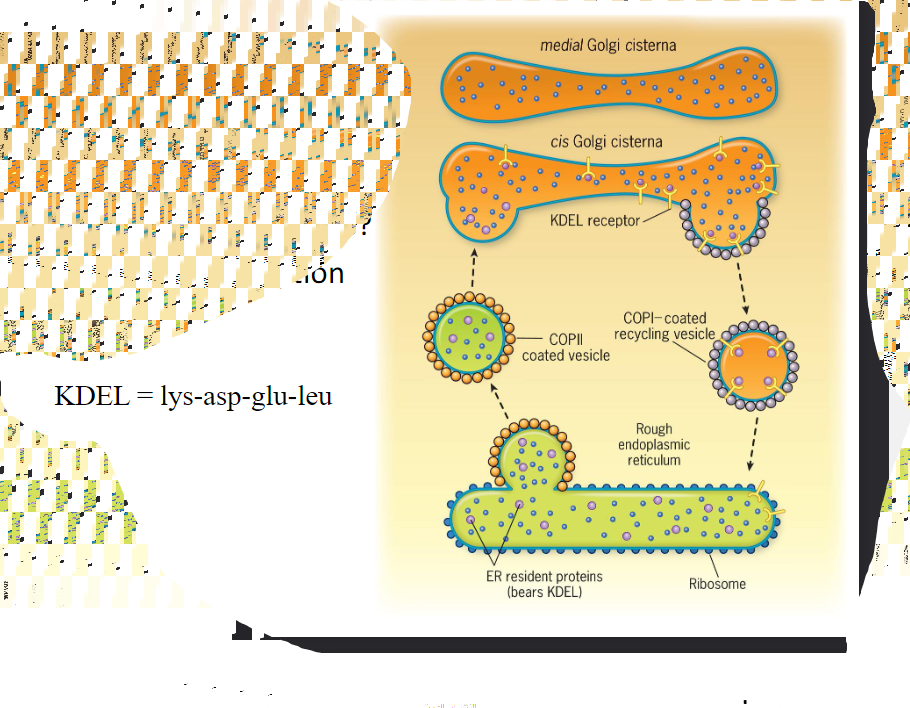
* Resident proteins of the ER have amino acid sequences that lead to their retrieval from the Golgi complex if theory is accidentally incorporated into a Golgi-bound transport vesicle. __Soluble ER proteins bear the retrieval signal KDEL__. Retrieval is accomplished as soluble ER proteins are going to bind to KDEL receptors that reside in the membranous wall of cis Golgi compartments. @@The KDEL receptors then bind to proteins of the COPI coat@@ allowing the whole complex to be recycled back to the ER>
* Look in book
* @@How does recognition occur?@@
* Resident proteins of the ER have amino acid sequences that lead to their retrieval from the Golgi complex if theory is accidentally incorporated into a Golgi-bound transport vesicle. __Soluble ER proteins bear the retrieval signal KDEL__. Retrieval is accomplished as soluble ER proteins are going to bind to KDEL receptors that reside in the membranous wall of cis Golgi compartments. @@The KDEL receptors then bind to proteins of the COPI coat@@ allowing the whole complex to be recycled back to the ER>
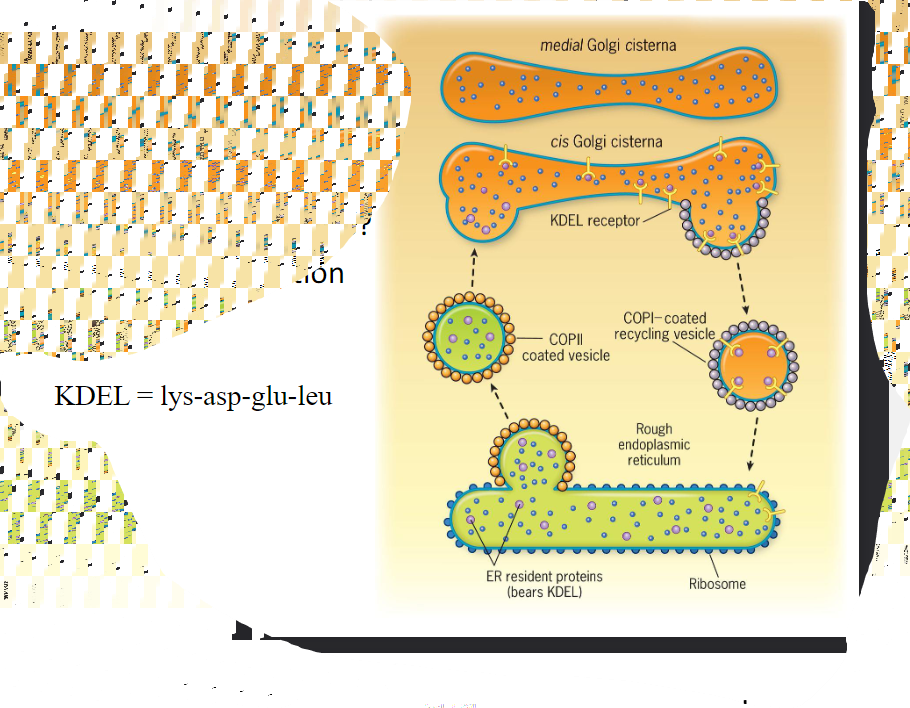
12
New cards
How are lysosomal enzymes targeted to the lysosome?
* Lysosomal enzymes are moved from the TGN in clathrin-coated vesicles and are targeted by cis cisternae that move a phosphorylated N-acetylglucosamine from nucleotide sugar donor to one or more mannose residues of N-linked oligosaccharides.
* How are they tagged for delivery to lysosomes?
* Mannose-6-phosphate
* How are they tagged for delivery to lysosomes?
* Mannose-6-phosphate
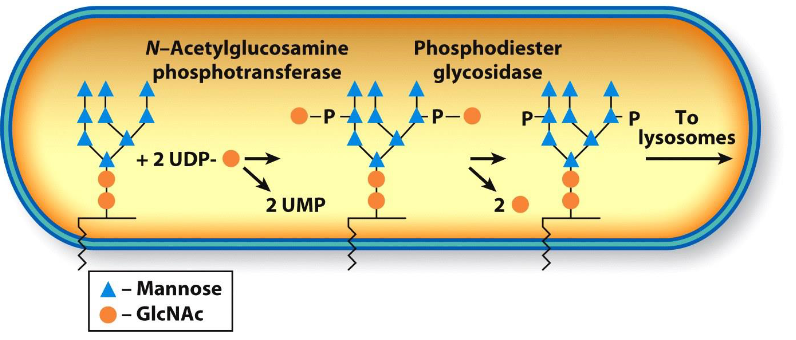
13
New cards
Lysosome formation
Look at picture:
1. cis Golgi cisternae
2. trans Golgi network
3. clathrin adaptor
4. dissociation of lysosomal enzyme from mannose 6-phosphate receptor / recycling of mannose 6-phosphate receptor
5. mannose 6-phosphate receptor (MPR)
6. endosome
7. endocytosis
1. cis Golgi cisternae
2. trans Golgi network
3. clathrin adaptor
4. dissociation of lysosomal enzyme from mannose 6-phosphate receptor / recycling of mannose 6-phosphate receptor
5. mannose 6-phosphate receptor (MPR)
6. endosome
7. endocytosis
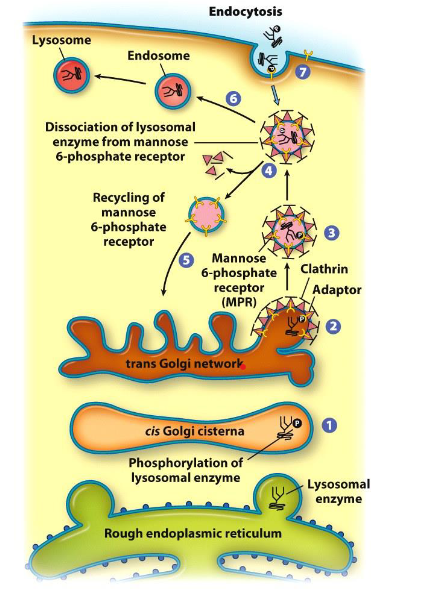
14
New cards
Describe in detail the 4 steps involved in the targeting of a vesicle to a particular membrane compartment and include important molecular components
1. Moving
2. Tethering
3. Docking
4. Fusing
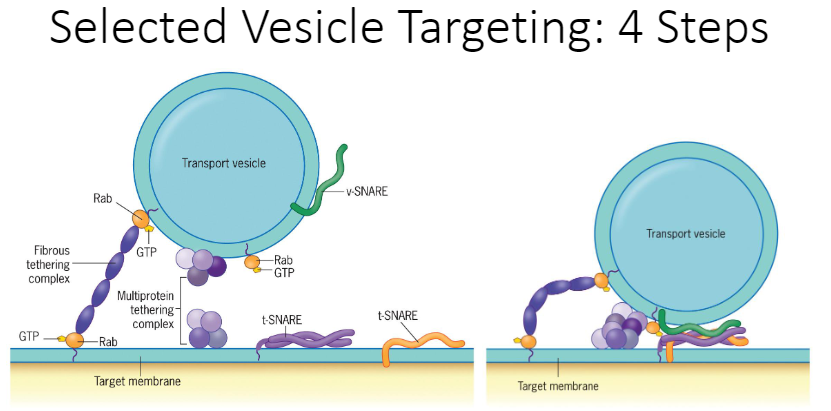
15
New cards
Moving
Moving is mediated by microtubules (microtubule transport) and motor proteins (motor protein transport)
16
New cards
Tethering
Is mediated by a collection of tethering proteins which can form a molecule bridge between 2 membranes. G-protein (Rab-GTP) is an on/off switch. There is a loose connection with the tether and target.
17
New cards
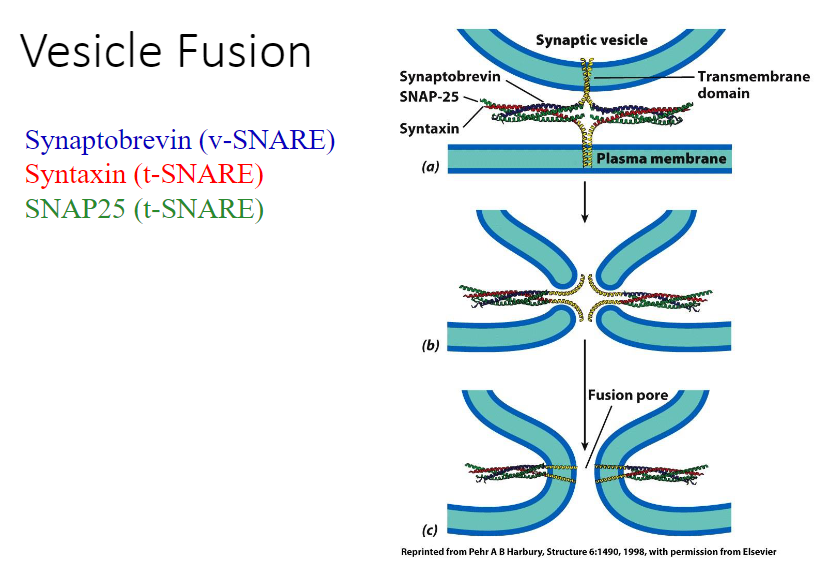
Docking
Has key proteins that make this possible (v-snares are for vesicles, t-snares are used for target proteins)
18
New cards
Fusing
The target and membrane compartment merge and form a connection. As the vesicle and site below fuses, there’s an opening between them; therefore, what was in the vesicle can get into the protein now.
19
New cards
How is the specificity of the process maintained?
Maintained through ^^exocytosis^^ where secretory vesicles go to the top of the membrane, and then are excreted
20
New cards
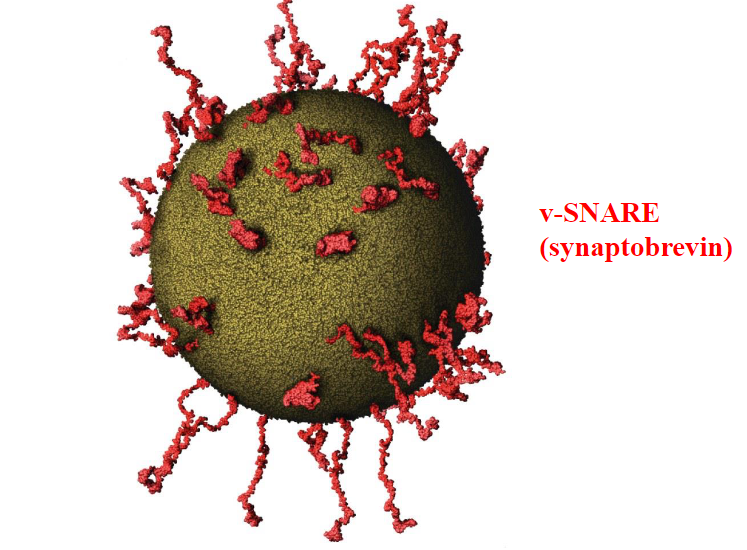
v-SNARE (synaptobrevin)
Involved in docking and fusion
21
New cards
Lysosome contents: Pick 2 enzymes and learn those
* Nucleases
* Acid ribonuclease; substrate: RNA
* Acid deoxyribonuclease; substrate: DNA
* Proteases
* Cathepsin; substrate: proteins
* Collagenase; substrate: collage
* Acid ribonuclease; substrate: RNA
* Acid deoxyribonuclease; substrate: DNA
* Proteases
* Cathepsin; substrate: proteins
* Collagenase; substrate: collage

22
New cards
Discuss the contents of a lysosome
1. Typically, a lysosome has __at least 50 different hydrolytic enzymes__ made in the RER and targeted to these organelles. Lysosomal enzymes can basically hydrolyze all biological macromolecules. All lysosomes have acidic enzymes called __acid hydrolases__ with a pH of 4.6.
2. The high internal proton concentration is maintained by a __proton pump in the lysosome’s boundary membrane.__
3. __Lysosomal membranes have a variety of highly glycosylated integral proteins with carbohydrate chains that form a protective lining and shield the membrane from attack by the enclosed enzymes.__
23
New cards
Define autophagy
* Lysosomes play a role in organelle turnover which is the regulated destruction and replacement of the cell’s own organelles.
* Definition
* During this process called __autophagy__: an organelle like mitochondria is surrounded by a double membrane structure/phagophore to make a double-membrane sequestering compartment called an autophagosome.
* Destruction and replacement of cellular organelles
* Hydrolytic enzymes will break apart those enzymes that have reached their limit
* Definition
* During this process called __autophagy__: an organelle like mitochondria is surrounded by a double membrane structure/phagophore to make a double-membrane sequestering compartment called an autophagosome.
* Destruction and replacement of cellular organelles
* Hydrolytic enzymes will break apart those enzymes that have reached their limit
24
New cards
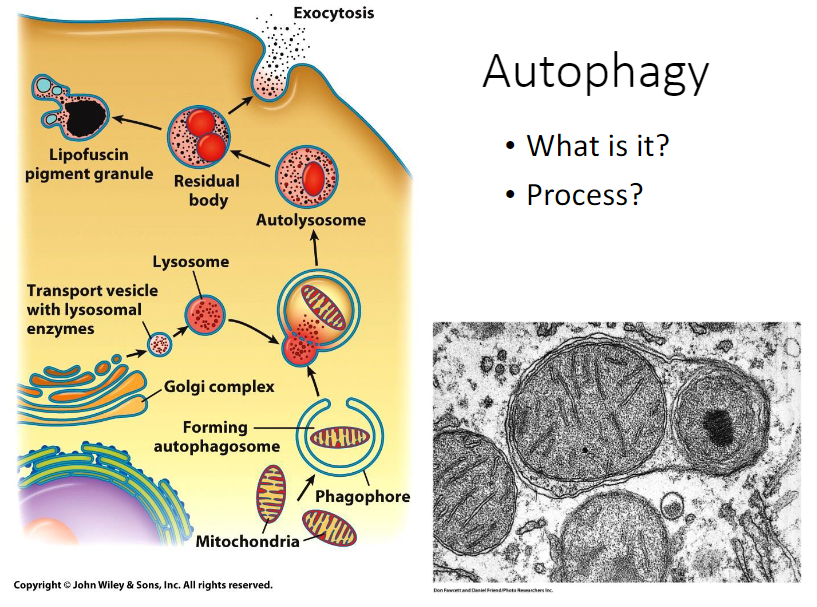
Autophagy process
1. In mammalian cells, __autophagosomes are thought to form de novo from contact sites between the ER and mitochondrial outer membrane.__
2. __Once formed, the outer membrane of the autophagosome fuses with a lysosome to make a structure called autolysosome.__
3. __After the digestive process of the autolysosome, the organelle is called a residual body.__
4. __Depending on the cell type, the residual body contents may be eliminated from the cell through exocytosis. Otherwise, they could be retained within the cytoplasm indefinitely as a lipofuscin granule.__
25
New cards
Phagocytosis
Carried out by a few types of cells specialized for the uptake of relatively large particles from the environment.
26
New cards
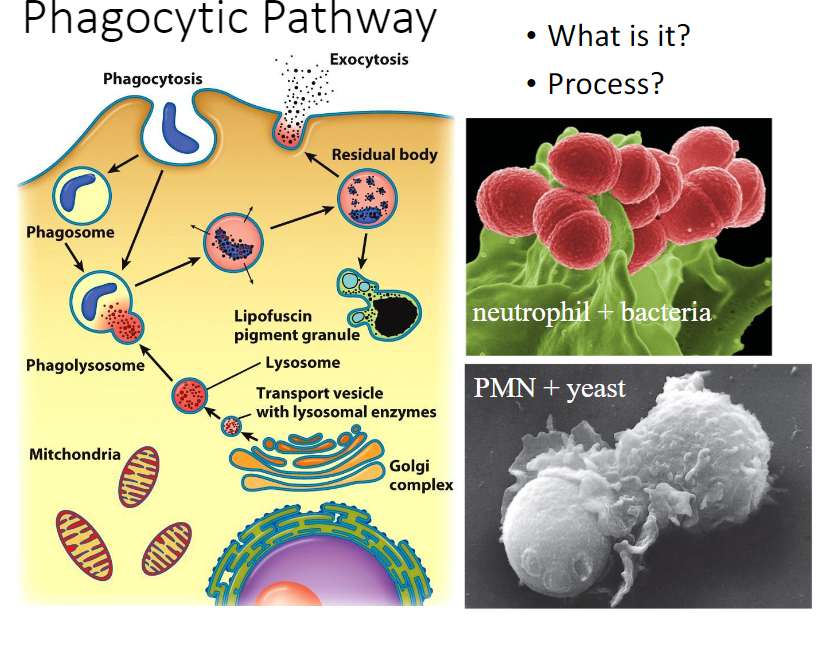
Phagocytic pathway
1. Professional __phagocytes__ like macrophages and neutrophils wander through the blood and tissues __phagocytizing invading organisms, damaged and dead cells, and debris. These materials are recognized and bound by receptors on the surface of the phagocyte prior to uptake.__
2. __Once inside the phagocyte, microorganisms may be killed by lysosomal enzymes or by oxygen-free radicals made within the lumen of the phagosome.__
3. __The engulfment of particulate material by phagocytosis is driven by contractile activities of the actin-containing microfilaments that underlie the plasma membrane.__
27
New cards
Defects in lysosomes
Tay-Sachs disease
28
New cards
What are the 2 basic mechanisms of endocytosis?
1. Bulk-phase endocytosis (pinocytosis)
2. Receptor-mediated endocytosis (RME aka clathrin-mediated endocytosis)
29
New cards
Bulk-phase endocytosis (pinocytosis)
1. __Definition__: Is the non-specific uptake of extracellular fluids
2. Any molecules (large/small) in the enclosed fluid can gain entry into the cell
3. Ex: Adding a substance to the culture medium like dye lucifer yellow/enzyme horseradish peroxidase, that is taken up by the cell nonspecifically.
4. Also removes portions of the plasma membrane and can function mostly in the recycling of the membrane between the cell surface and interior compartments
30
New cards
Receptor-mediated endocytosis (RME aka clathrin-mediated endocytosis)
__Definition__: Brings about the uptake of specific extracellular macromolecules (ligands) following their binding to receptors on the external surface of the plasma membrane
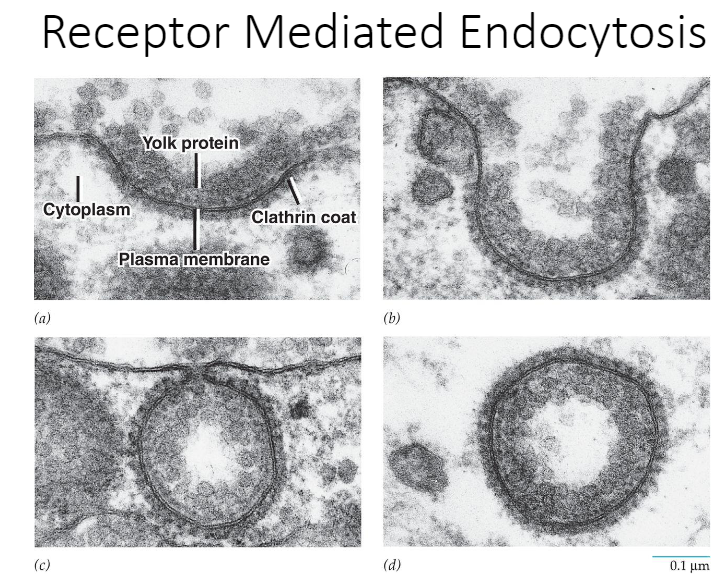
31
New cards
Similarities between bulk-phase endocytosis and receptor-mediated endocytosis
* __Both decrease the plasma membrane surface area__
32
New cards
COP2 vs clathrin similarities
* G-proteins
* Receptors
* Adaptors
* Coat protein
* Receptors
* Adaptors
* Coat protein
33
New cards
Structure of clathrin-coated vesicles in RME
1. __Clathrin triskelion (light chain and heavy chain [N-terminal hook]) and Adaptor protein (AP2) complex__
2. Arrangement of triskelions and adaptors in the outer clathrin coat.
3. The sides of the polygons are made by parts of the legs of the overlapping triskelions.
34
New cards
Clathrin-triskelion and AP2 adaptor roles
1. __The N-terminus of each clathrin-heavy chain makes a “hook” that projects toward the surface of the membrane where it engages an adaptor.__
2. Each adaptor, which consists of 4 different polypeptide subunits, can bind a diverse array of accessory proteins.
3. Both the hooks and the adaptors are at the vertices of the polyhedrons.
4. **Formation of clathrin coated vesicle**
* **Clathrin triskelion**
* **AP2 adaptor connects clathrin to the membrane**
35
New cards
Clathrin vesicles: role of dynamin
* Dynamin helps pinch off the vesicle
* Dynamin wraps itself on the membrane that is left, squeezes it and pinches it off, GTP hydrolysis
* Dynamin wraps itself on the membrane that is left, squeezes it and pinches it off, GTP hydrolysis
36
New cards
Function of dynamin and GTP
1. __Dynamin is a large GTP-binding protein required for the fission of the vesicle from the membrane on which it forms__
__Summary of steps__: Dynamin subunits, clathrin lattice, dynamin ring, GTP hydrolysis or GTPyS
37
New cards
Function of dynamin and GTP: step 0
The clathrin lattice of the coated pit
38
New cards
Function of dynamin and GTP: step 1
Undergoes rearrangement to make an invaginated vesicle connected to the overlying plasma membrane by a stalk
39
New cards
Function of dynamin and GTP: step 2
Th dynamin subunits concentration the region undergoes polymerization to make a ring around the stalk
40
New cards
Function of dynamin and GTP: step 3
Changes in the conformation of the ring, which are thought to be induced by GTP hydrolysis
41
New cards
Function of dynamin and GTP: step 4
Lead to fission of the coated vesicle from the plasma membrane and disassembly of the dynamin ring
42
New cards
Function of dynamin and GTP: step 5
If vesicle budding occurs in the presence of GTPyS, a nonhydrolyzable analog of GTP, dynamin polymerization continues beyond the formation of a simple collar, producing a narrow tubule constructed from several turns of the dynamin helix
43
New cards
Discuss, in general, the endocytic pathway paying attention to the roles played by early endosomes, late endosomes, and the sorting compartment (figure 8.45).
1. Receptors taken up by endocytosis are transported in vesicles to __early endosomes which serve as a sorting station__ that direction different types of receptors and ligands along different paths
2. Pathway:
1. The movement of materials from the extracellular space to early endosomes where sorting occurs
2. Endocytosis of two types of receptor-ligand complexes:
1. LDL receptors are typically sent back to the plasma membrane
2. Ligands are transferred to late endosomes
3. __Signaling receptors like the EGF receptor is typically transported to late endosomes along with their ligands__.
* __Late endosomes__ also receive newly synthesized lysosomal enzymes from the TGN. These enzymes are carried by mannose 6-phosphate receptors (MPRs), which return to the TGN. The contents of late endosomes are transferred to lysosomes by a number of routes.
* Early endosomes
* Change pH, sorting compartment (receptors that go back to the surface vs other receptors that aren’t), the molecules that are taking in go further, eventually, it gets to the late endosomes
* Endosomes
* Membrane compartments
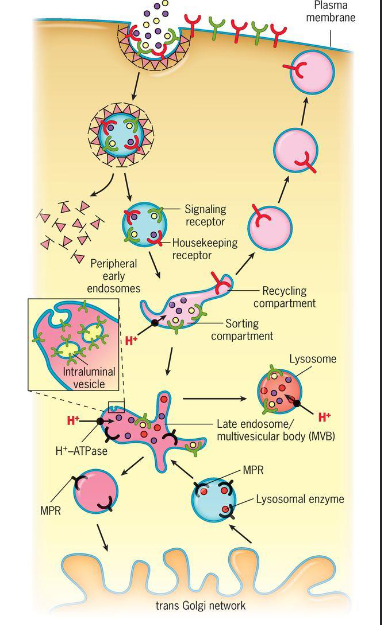
44
New cards
Housekeeping receptors
Receptors taken up by endocytosis are transported to an early endome which serves as a sorting station that directs the different types of receptors and ligands along different pathways. __Housekeeping receptors__ typically disassociate from their bound ligands as a result of the high H+ concentration of the early endosomes. The receptors are then concentrated into specialized tubular compartments of the early endosome which represent recycling centers. Vesicles that bud from these tubules curry receptors back o the plasma membrane for additional rounds of endocytosis.
45
New cards
Signal receptors
In contrast, __released ligands__ become concentrated into a sorting compartment before being dispatched to a late endosome and ultimately to a lysosome, where final processing occurs.
46
New cards
Housekeeping vs signaling receptors
Taking in things into the cell that the cell wants to take in, the red ones go back to the surface again, the early endosome sorts the housekeeping ones so that they stay in the surface?; the stuff in the sorting station that didn’t get sent out go to the sorting endosomes
47
New cards
Compare in general terms the way proteins are transported into the RER with how they are transported into the nucleus, peroxisomes, mitochondria, and chloroplasts. Explain, in detail, how proteins enter the mitochondria. Be sure to delineate between membrane and matrix proteins.
1. Nucleus
1. Book says will be discussed in a later section
2. Peroxisomes
1. Able to import peroxisomal matrix proteins in their native, folded conformation.
3. Mitochondria and chloroplasts
1. Both import proteins that must assume an unfolded state.
2. Chloroplasts
1. The majority of proteins are imported from the cytosol. The outer and inner envelope membranes have distinct translocation complexes that work together during import. Chaperones aid in the unfolding of the polypeptides in the cytosol and the folding of the proteins in the chloroplast. Most proteins are synthesized with a removable N-terminal sequence (called transit peptide) highly variable in length and sequence.
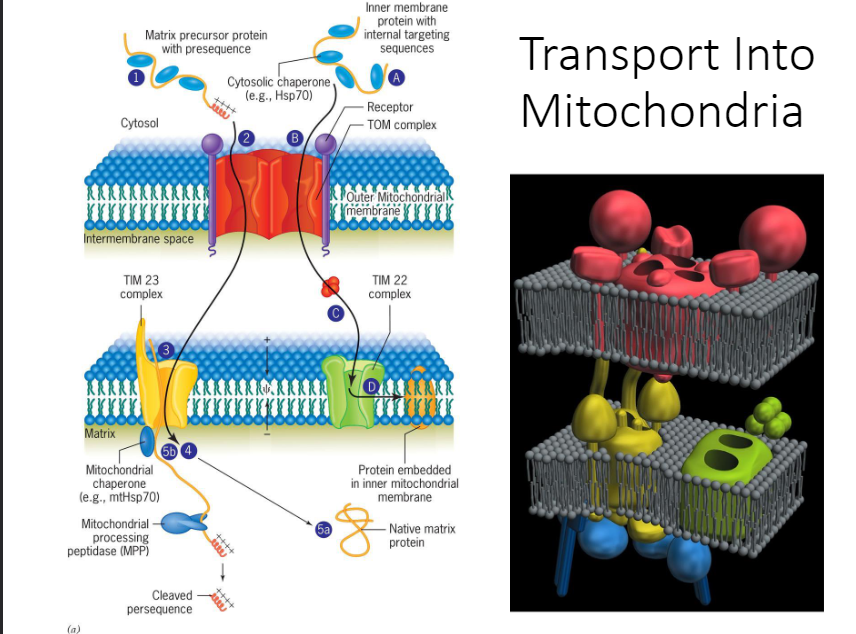
4. How proteins enter the mitochondria
1. Step 1: Proteins are imported posttranslationally into either the mitochondrial matrix/the inner mitochondrial membrane. The polypeptide is targeted to a mitochondrion sequence located at the N-terminus in the matrix protein
2. Step A: And is located internally in the most inner membrane proteins
3. Step 2/B: Cytosolic Hsp70 molecules unfold the polypeptides prior to their entry into the mitochondrion. The proteins are recognized by membrane receptors and translocated through the OMM by way of pores in the TOM complex of the OMM.
4. Step C: Most integral proteins of the IMM are the direction of the TIM22 complex of the IMM
5. Step D: Which steers them into the lipid bilayer of the IMM.
6. Step 3: Mitochondrial matrix proteins are translocated through the TIM23 complex of the IMM.
7. Step 4: Once the protein enters the matrix, it’s bound by a mitochondrial chaperone
8. Step 5a: Which may either pull the polypeptide into the matrix or act like a Brownian ratchet to ensure that it diffuses into the matrix. Once in the matrix, the unfolded protein assumes its native conformation with the help of Hsp60 chaperones.
9. Step 5b: The presequence is removed enzymatically.
48
New cards
Peroxisomes
Able to import peroxisomal matrix proteins in their native, folded conformation.
49
New cards
Mitochondria and chloroplasts
1. Both import proteins that must assume an unfolded state.
2. Chloroplasts
1. The majority of proteins are imported from the cytosol. The outer and inner envelope membranes have distinct translocation complexes that work together during import. Chaperones aid in the unfolding of the polypeptides in the cytosol and the folding of the proteins in the chloroplast. Most proteins are synthesized with a removable N-terminal sequence (called transit peptide) highly variable in length and sequence.
50
New cards
How proteins enter the mitochondria
* Fully made protein has to be denaturated (chaperones help keep it open/unfolded; it can’t get in if its folded)
* TOM (transfer outer membrane) complex gets it to pass through the outer membrane
* Then the proteins go through a TIN (transfer inner membrane) complex; TIN 23 (comes through the TOM, enters the TIM and go to the matrix) and TIN 22 (some proteins stay in the membrane like ATP synthase/ETC; allows it to stay in the membrane; membrane embedded proteins)
* TOM (transfer outer membrane) complex gets it to pass through the outer membrane
* Then the proteins go through a TIN (transfer inner membrane) complex; TIN 23 (comes through the TOM, enters the TIM and go to the matrix) and TIN 22 (some proteins stay in the membrane like ATP synthase/ETC; allows it to stay in the membrane; membrane embedded proteins)