Biology - DNA (No Biotech)
1/97
Earn XP
Description and Tags
Nathan Sewart xx
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
98 Terms
Chromosomes are (state)
made of a very long strand of helical DNA
Locus of a gene is (state)
the location of a gene on a chromosome
Chromatin is (state)
long and thin tangled strands of DNA (when the cell is not dividing)
Chromatids are (state)
condensed, shorter, thicker strands of DNA (during cell division).
Compare chromosomes in prokaryotes and eukaryotes.
Prokaryote DNA (including mitochondria and chloroplasts) is circular, unbound, found in the cytosol and has no introns. Eukaryotic DNA is linear, bound to histones, found in the nucleus, and has introns and exons.
Describe the structural properties of the DNA molecule, including: nucleotide composition and pairing (6 Responses)
DNA is a helical double-stranded molecule. It runs in a 5' to 3' direction.
It is made of a nucleotides. Said nucleotide has a nitrogenous base (A T C G) that is attached to a deoxyribose sugar. Said sugar is linked to a phosphate group.
The phosphate group from each nucleotide bonds to the deoxyribose sugar on the next nucleotide to form a sugar-phosphate backbone.
The length of nucleotide bonded to another nucleotide via a weak hydrogen bond is roughly 2 nano meters.
Adenine complementary base pairs with Thymine. Cytosine complementary base pairs with Guanine.
Phosphate group gives DNA an overall negative charge.
Describe the structural properties of the DNA molecule, including: the weak bonds between strands of DNA that allow for replication.
Weak hydrogen bonds form between complementary bases. Hydrogen bonds hold the two strands of DNA together and are weak enough to be broken during transcription and DNA replication.
Explain the importance of complementary base pairing (A–T and C–G).
Allows the DNA molecule to be replicated. This is because if the sequence of one strand is known, the sequence of the complementary strand is also known. Therefore, each strand of DNA can be used as a template to synthesis another complementary strand of DNA.
Describe and represent the process of semi-conservative replication of DNA (5 Steps)
1. DNA unwinds and DNA helicase breaks weak hydrogen bonds between bases, splitting the DNA strands.
2. Each strand serves as a template for the making of a new strand.
3. Free DNA nucleotides link into position and complementary base pair according to A-T and C-G.
4. DNA polymerase links the nucleotides to form new strands and Ligase seals the sugar-phosphate backbone.
5. Two new DNA molecules created which are replicas of the original. This is "semi conservative" because each strand contains 1 old and 1 new strand.
A gene is (state)
a section of DNA which codes for a RNA molecule.
Ribonucleic acid is (3 Features and name types)
Single stranded.
Has Uracil bases instead of Adenine.
Has ribose sugar.
Three types: mRNA, tRNA, and rRNA.
Distinguish between exons and introns as coding and non-coding segments of DNA found in genes in eukaryotes. (State both, and 1 key concept)
Exons are the DNA in a gene transcribed into a molecule of messenger RNA and then translated into a polypeptide.
Introns are the DNA in a gene that is only transcribed into an RNA molecule. It does not code for proteins.
Exon coding / Intron non-coding
Prokaryote cell's DNA does not contain Introns.
Describe how both exons and introns are transcribed but only the information contained in exons is translated to form a polypeptide in eukaryotes (3 steps, outcome for exon and intron)
1. Post-transcriptional processing splices out introns.
2. Introns remain in the nucleus. Exons are kept in the mRNA and exit the nucleus.
3. After introns are spliced out, the remaining exons are joined together by ligase to form a shorter mRNA molecule, which is then translated.
4. Therefore, both exons and introns are transcribed but only the information contained in exons is translated to form a polypeptide.
5. Introns code for ribosomal RNA (rRNA, involved in translation), transfer RNA (tRNA, involved in translation), micro RNA (miRNA, involved in regulation of gene expression).
Protein synthesis consists
of transcription and translation.
Protein synthesis occurs in (prok)
the cytosol in prokaryotes
Describe and illustrate the role of DNA, mRNA, transfer RNA (tRNA), and ribosomal RNA (rRNA) in transcription and translation. DNA
Located in the nucleus, contains genes that are hereditary units. The template strand provides the code to synthesise a molecule of mRNA.
Describe and illustrate the role of DNA, mRNA, transfer RNA (tRNA), and ribosomal RNA (rRNA) in transcription and translation. mRNA
Located in the nucleus, cytosol and ribosomes. Is single stranded. mRNA is a complimentary copy of the DNA template strand and carries the code to the ribosomes in the cytoplasm.
Describe and illustrate the role of DNA, mRNA, transfer RNA (tRNA), and ribosomal RNA (rRNA) in transcription and translation. tRNA
Located in cytoplasm and ribosomes. Single strand, shaped like a 3D clover. About 80 nucleotides long. Carries an amino acid during translation. One end of the tRNA attaches to a specific amino acid and the other end has anticodon which attaches to a codon on the mRNA. Each tRNA can only carry one specific amino acid.
Describe and illustrate the role of DNA, mRNA, transfer RNA (tRNA), and ribosomal RNA (rRNA) in transcription and translation. rRNA
Ribosomes are made up of rRNA and protein molecules. Ribosomes have a large subunit and small subunit. Ribosomes move along the mRNA, translating the mRNA code into a sequence of amino acids in a polypeptide.
Describe and illustrate the role of DNA, mRNA, transfer RNA (tRNA), and ribosomal RNA (rRNA) in transcription and translation. miRNA
Located in the nucleus. About 22 nucleotides long. Believed to regulate gene expression after transcription. Bind to mRNA molecules and prevent translation of these sections.
Transcription is the
Process of a DNA molecule being read and rewritten into a mRNA molecule.
Transcription occurs (euk and prok)
in the nucleus of eukaryotic cells and in nucleoid region of prokaryotic cells.
State the steps of transcription (3)
1. DNA unzips and unwinds as the hydrogen bonds are broken by helicase.
2. Free floating RNA nucleotides attach to the exposed DNA bases on template strand with the assistance of RNA polymerase.
3. mRNA strand unzips from the DNA template strand with help of RNA polymerase. DNA rewinds.
State the steps of Post transcriptional processing (2)
1. Introns spliced out and exons joined via ligase enzyme.
2. Mature mRNA leaves nucleus (eukaryotes) and moves to ribosome.
Introns and exons allow
multiple proteins to be synthesised from a single mRNA, increasing complexity of an organism. Most prokaryotes do not have introns, meaning their entire length of DNA codes for proteins.
State the steps of translation (3)
1. Initiation: Ribosome binds to mRNA strand at start codon.
2. Chain elongation: tRNA brings specific amino acid to ribosomes, where its anticodon binds with complementary mRNA codon. Ribosome moves along to next codon, as tRNA brings next amino acid. Peptide bonds join amino acids.
3. Termination: Stop codon stops protein synthesis, as mRNA disintegrates. Polypeptide chain created.
Translation is the
Process of a polypeptide chain being built from a codon sequence on the mRNA molecule.
Translation occurs in
the cytoplasm.
hard state all steps of protein synthesis (8)
1. DNA unzips and unwinds as the hydrogen bonds are broken by helicase.
2. Free floating RNA nucleotides attach to the exposed DNA bases on template strand with the assistance of RNA polymerase.
3. mRNA strand unzips from the DNA template strand with help of RNA polymerase. DNA rewinds.
4. Introns spliced out and exons joined via ligase enzyme.
5. Mature mRNA leaves nucleus (eukaryotes) and moves to ribosome.
6. Initiation: Ribosome binds to mRNA strand at start codon.
7. Chain elongation: tRNA brings specific amino acid to ribosomes, where its anticodon binds with complementary mRNA codon. Ribosome moves along to next codon, as tRNA brings next amino acid. Peptide bonds join amino acids.
8. Termination: Stop codon stops protein synthesis, as mRNA disintegrates. Polypeptide chain created
Describe the relationship between DNA codons, RNA codons, anticodons, and amino acids. (state the 6 forms, from start to finish)
(T) ↓ DNA Complementary Strand:
(T) ↓ DNA Template Strand:
(U) ↓ mRNA (codon):
(U) ↓ tRNA (anticodon):
(Aa) ↓ Amino Acids:
(Pp) ↓ Polypeptide:
Codon is a
Sequence of 3 nucleotides.
DNA Complementary Strand: GGA AAT CCA TTA
State tRNA
CCU UUA GGU AAU
For Codon questions, DNA can be
read up and down.
i.e: ATT OR TTA
Why 3 bases in Codon?
Sequence of 3 bases that corresponds to an amino acid. As there are 4 bases, a total of 64 different codons. However, if codon were 2 bases, only 16 different codons. So, 64 bases needed.
Template strand is used to
translate the mRNA. It is complementary to mRNA strand.
Coding strand contains the
exact same sequence of nucleotides as the mRNA (except U for T). It is not used as template for transcription.
Distinguish between coding (gene) and template strands of DNA (2 key ideas, mostly asked in multi choice).
Template strand is used to transcript the mRNA. It is complementary to mRNA strand.
Coding strand contains the exact same sequence of nucleotides as the mRNA (except U for T). It is not used as template for transcription.
Recognise that DNA strands are directional and are read 5’ to 3’.
"Read up, write down".
DNA Polymerase will read a strand 3' to 5', and Reads: 3' – 5', Writes: 5' – 3'.
The folding of a polypeptide to form a protein with a unique three-dimensional shape is determined by
its sequence of amino acids.
Describe the factors that determine the primary, secondary, tertiary, and quaternary structure of proteins. (4 Steps)
1. Primary structure is an amino acids polypeptide chain linked. DNA sequence determines mRNA sequence which determines amino acid sequence.
2. Secondary: Folding of polypeptide chain due to hydrogen bonds form, forming an alpha helix or beta sheet.
3. Tertiary: amino acids form bonds, causing the polypeptide to fold into a complex 3D structure. Number, type, and sequence of amino acid determines the shape of protein. This determines function.
4. Quaternary: two or more polypeptide chains bind together; not all proteins have quaternary structure
Proteins are essential to
cell structure and function.
Examples of proteins with specific shapes include (4 Answers)
enzymes, some hormones, receptor proteins, and antibodies.
Proteins have a unique shape determined by
protein folding.
Explain why the three-dimensional shape of a protein is critical to its function. Hormones
For hormones, they are chemical messengers which work by binding to receptors. If its shape changes it can't bind to its receptor and thus loses its function.
Explain why the three-dimensional shape of a protein is critical to its function. Receptors
For receptor proteins, they work by having complementary shape to a messenger molecule, initiating a cellular response when they bind. If its shape changes, it can't bind to its messenger and no cellular response.
Explain why the three-dimensional shape of a protein is critical to its function. Antibodies
For antibodies, they have unique shape to a complementary antigen, allowing the immune system to recognise an antigen as self or foreign. If the shape changes, it cannot recognise the antigen and may potentially lead to infection. Also, if an its shape changes into one complementary to a self-antigen, can lead to autoimmune disease.
Explain why the three-dimensional shape of a protein is critical to its function. Enzymes
For enzymes, they work by catalysing reactions by binding to substrate molecules with complementary shape. If the shape changes, it cannot bind and thus loses its function.
Enzymes are specific for
their substrate
Enzymes increase
reaction rates by lowering activation energy.
Enzymes catalyse _________ by ____________
metabolic reactions in cells by binding to substrates.
The active site of the enzyme binds to
a substrate
Enzymes are (6)
globular proteins
have a specific shape and active site
are reused
increase the rate of reaction and speed up chemical reactions
provide an alternative pathway with a lower activation energy
Enzymes are not used up in reactions.
Exergonic reaction
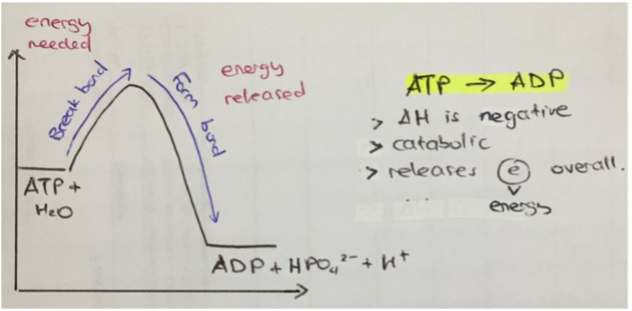
releases energy (exit). Usually catabolic, (as energy is released from broken bonds within a molecule). Usually occurs when the reactants (i.e., ATP) contain more energy than the products (i.e., ADP + Pi). For example (see image), as bonds are broken (until the hump), the amount of "free energy" increases, as breaking bonds releases energy. As bonds form (after the hump), the amount of "free energy" decreases, as synthesising bonds absorbs energy. However, the total amount of free energy decreased, meaning energy was released (can be used in the cell)
Endergonic reaction
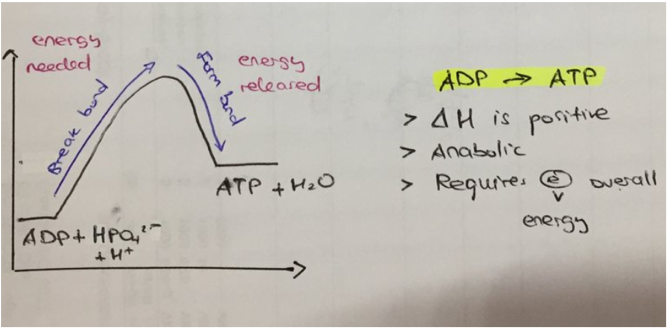
absorb energy (enter). Usually anabolic, ((building up), as energy is required to synthesise bonds between molecules). Usually occurs when the reactants (i.e., ADP + Pi) contain less energy than the products (i.e., ATP). For example (see image), as bonds are broken (until the hump), the amount of free energy increases, as breaking bonds releases energy. As bonds form, the amount of free energy decreases. However, the free energy does not decrease more than the initial amount, meaning the total free energy increased. This means energy was absorbed (can be broken for future use).
Initial energy input needed for reactions is called
activation energy. In graphs, seen as the initial hump of energy. Enzymes lower this hump (see image).
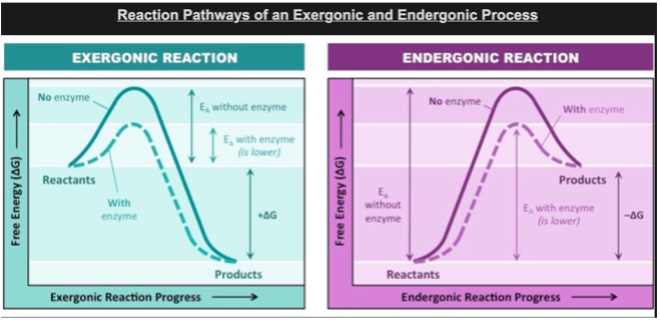
Enzymes increase reaction rate by lowering activation energy by: (2)
bringing substrate molecules together in correct orientation or places stress on substrate bonds leading them to break more easily.
Describe the induced-fit model of enzyme–substrate binding.
Substrate binds complementary to active site of enzyme. Shape of shape of enzyme and substrate make a conformational change for induced fit. Bonds of substrate are strained. Products released and enzyme returns to original shape. Enzyme is reused.
Substrate (Define)
molecules that bind to enzymes.
Active site (define)
site of enzyme that binds to substrate.
Substrates must bind for
enzymes to function.
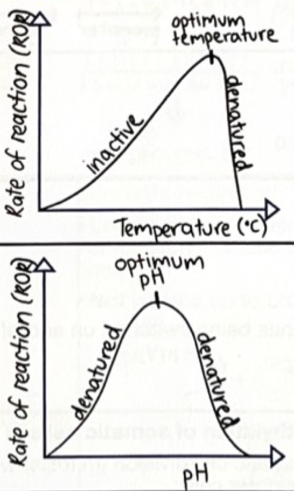
Enzymes have specific functions and are affected by factors including: Temperature (2 responses)
Enzymes have optimum temperature.
High temperature: enzyme denatures. Shape of active site changes, meaning substrate cannot bind. Denaturation is irreversible.
Low temperature: slows enzymes until inactive as there is not enough kinetic energy for enzyme to collide with substrate. Reversible.
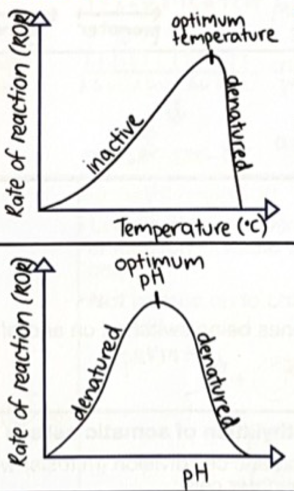
Enzymes have specific functions and are affected by factors including: pH (1 response)
Enzymes have optimum ph.
High or Low pH: enzyme denatures. Shape of active site changes, meaning substrate cannot bind. Denaturation is irreversible.
Enzymes have specific functions and are affected by factors including: presence of inhibitors (2 responses)
Competitive Inhibitors: has similar shape to substrate. Competes with substrate binding to the active site of enzyme, leading to substrate not binding. Reversible.
Non-competitive Inhibitors: Binds to place on enzyme other than the active site. Changes shape of enzyme active site, meaning substrate cannot bind. Usually, irreversible.
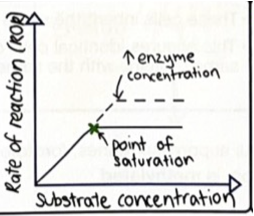
Explain how The rate of an enzyme-controlled reaction is affected by: concentrations of reactants.
As substrate concentration increases, rate of reaction increases to a point. Plateaus as all enzyme active sites are full.
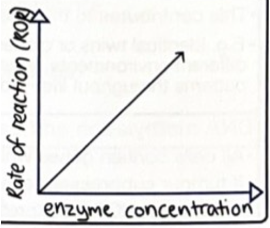
The rate of an enzyme-controlled reaction is affected by: concentration of the enzyme.
As enzyme concentration increases rate of reaction increases. This increase in rate of reaction is until substrate concentration becomes a limiting factor.
The phenotypic expression of genes depends on
factors controlling transcription and translation. These include the products of other genes, such as transcription factors, and the environment.
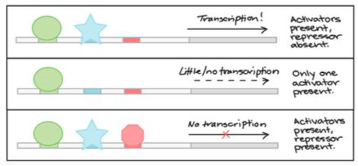
Transcription factors __________________________________
This ______________________
Activator proteins _____________
while repressor proteins ________
activate transcription of a specific gene by binding to gene's promoter sequence.
This recruits other factors and RNA polymerase. Activator proteins increase rate of transcription while repressor proteins inhibit it
Gene (define)
section of DNA which contains information for a specific function, for example coding for a protein.
Gene expression (define)
when different combinations of genes are expressed and repressed.
Cell differentiation is when
Cells express specific genes while repressing others to perform their function. Process by which cells specialise.
Stem cell (is a)
unspecialised cell.
Cellular differentiation associated with tissue growth and development is controlled by
gene expression.
Cellular differentiation results in
stem cells becoming specialised.
Occurs as genes get switched on and off due to methylation. Examples include nerve and muscle.
Recognise that changes in DNA methylation and histone modification can alter gene expression.
DNA is negatively charged and wraps around positively charged histone proteins. DNA which wraps too much is not accessible for transcription.
Methyl group attaches to cytosine nucleotide on promotor region of a gene. Means transcription factor cannot bind, so gene is switched off, so gene is not expressed, so no protein.
Most commonly methylated base is
Cytosine
Epigenetic changes can lead to
phenotypic differences between identical siblings, phenotypic differences between clones, and may cause human diseases
Genotype (define)
genetic make-up of an organism.
Phenotype (define)
genotype + environmental influences. Is physical appearance.
Phenotypic expression (define)
relates to physical, biochemical, physiological characteristics
Epigenetic changes …
does not change DNA sequence but affects gene expression. It determines whether genes are on.
Usually, it is methylation in genes and can be caused by environmental factors. These are random. Contributes to the phenotype of the individual. I.e., identical twins or clones have the same DNA, but different environments, leading to different methylation patterns throughout life, and thus different genes expressed.
Explain the relationship between Mitosis and Methylation:
Somatic cell division (mitosis) will produce identical daughter cells. These cells inherit the same gene methylation patter. This ensures identical cells continue to produce the same proteins with the same functions.
Explain how epigenetic modifications in genes that control cell division, such as changes in DNA methylation, can lead to cancer.
_____________ that _______ can cause _________, _______. All ______ contain ______ that _____.
State the three types of genes involved.
Epigenetic modifications in genes that control cell division can cause cells to divide uncontrollably, leading to cancer. All cells contain genes that control cell division.
Tumour suppressor genes (if methylated -> Cancer)
Protooncogenes (promotes cell cycle, if methylated -> Cancer),
Oncogenes (promote cancer, if not methylated -> Cancer).
Summarise methylation:
A gene is switched on if unmethylated, meaning acetylated histones (DNA loosely packed). This means the gene is active and expressed. Gene is off when methylated and deacetylated histones (DNA tightly packed). Gene is silent and not expressed.
State the genes involved in cancer (3)
Tumour suppressor genes (if methylated -> Cancer)
Protooncogenes (promotes cell cycle, if methylated -> Cancer),
Oncogenes (promote cancer, if not methylated -> Cancer).
Changes in the DNA sequence are called
mutations
Mutation (define)
any change in nucleotide sequence. Generally, a permanent change in the DNA.
What is the result of a mutation?
Change in DNA sequence leads to change of mRNA, leads to change in structure/shape of proteins.
Mutations in genes and chromosomes can result from (2 responses).
errors in DNA replication or cell division
damage by physical or chemical factors in the environment.
Types of DNA Mutations (3 responses)
Substitution: a base is replaced with another.
Insertion: Insertion of one or more bases into the DNA.
Deletion: The deletion of one or more bases.
Types of Chromosome Mutations (6 respones, not in exam just keep in mind).
Deletion: Deletion of a whole gene (green).
Inversion: A region of DNA switches ends (yellow).
Reciprocal Translocation: Two non-homologous chromosomes exchange genetic material (pink).
Robertsonian Translocation: Two chromosomes join and act as one.
Duplication: section of DNA is abnormally copied (blue).
Genome mutation: change in the number of chromosomes.
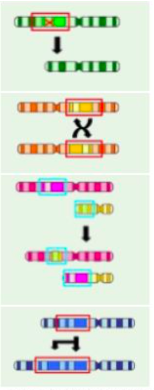
A Mutagen is (define)
factor that increases rate of mutation.
Mutation rate can be increased by:
o ionising radiation
o mutagenic chemicals
o viruses.
Name the types of ionising radiation that increase mutation rate (3 responses)
UV
X-rays
Gamma radiation.
Name the types of mutagenic chemicals that increase the mutation rate (2 responses)
Smoking
Bleach.
Name the types of viruses that increase mutation rate (1 response)
Viruses
Compare the different potential consequences of mutations in germ cells and somatic cells (2 responses).
If a mutation occurs in a germ cell, every cell in the organism will carry the mutation. Some of the gametes will carry mutation, potentially passing it on to children.
If a mutation occurs in a somatic cell, it is localised in the body. Not passed on to children, so no gametes carry mutation.
Explain how inheritable mutations can lead to changes in the characteristics of the descendants.
If a germline mutation can result in deleterious, neutral, or advantageous mutations. I.e., unfavourable traits, making it less likely to survive, no effect, or gives an organism favourable trait, making it more likely to survive.