2.3: Vesicular Traffic to and From the Cell Surface
1/44
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
45 Terms
Secretion of Insulin in Pancreatic β-Cells: High Glucose
when blood glucose levels rise, β-cells take up glucose
glucose metabolism increases ATP production
a high ATP/ADP ratio closes ATP-sensitive K+ channels → depolarization → Ca2+ influx
Ca2+ triggers exocytosis of insulin-containing secretory granules

Secretion of Insulin in Pancreatic β-Cells: Retrograde Retrieval
pancreatic β-cells package proinsulin → insulin into vesicles that bud from the trans Golgi network (TGN)
the early vesicles are called immature secretory granules
when the vesicle first buds off, it contains extra “stuff” (insulin/proinsulin, membrane proteins, processing enzymes, etc.)
retrograde retrieval: as vesicles mature, the cell retrieves unwanted proteins back to the Golgi (vesicle → Golgi)
as extra proteins leave, insulin becomes more concentrated
the dense mature granules can rapidly release lots of insulin
they dock at the PM (inner surface) and are primed for rapid release (exocytosis)
Clathrin Vesicles
bud from PM → endosomes (endocytosis)
trans-Golgi network → endosomes/lysosomes
specialize in cargo selection
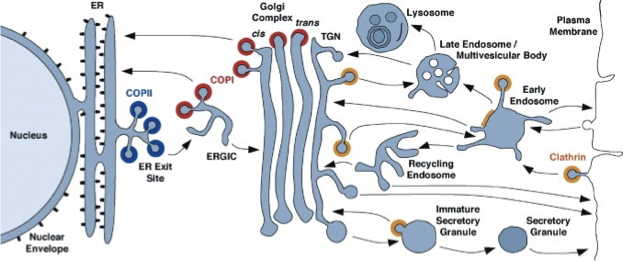
All Membrane-Bound Compartments are Connected
proteins are successively modified as they pass thru a series of compartments
some vesicles select cargo molecules and move them to the next compartment
some vesicles retrieve escaped proteins and return them to a previous compartment
the biosynthetic secretory is a continuous flow of material
Big Picture of Vesicles
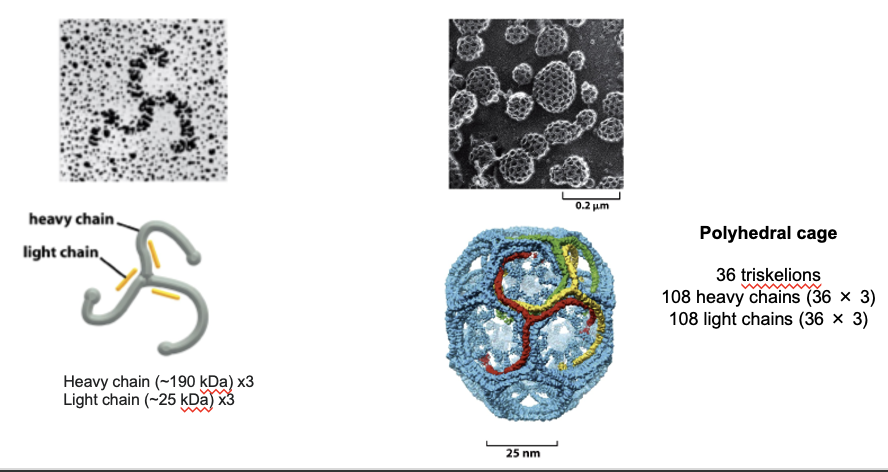
Clathrin Triskelion
clathrin is the main structural protein that forms the outer shell of the vesicle during endocytosis or TGN sorting
forms a 3-legged structure (triskelion): has 3 heavy and 3 light chains
the shape allows it to link to other triskelions at flexible joints
isolated triskelions spontaneously self-assemble into a polyhedral cage
the cage bends the membrane and gives vesicles their shape
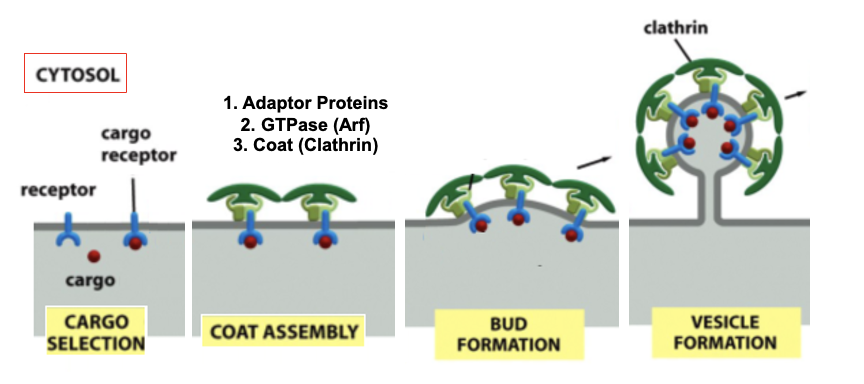
Clathrin Vesicle Formation: Step 1
adaptor proteins (AP) form a discrete second layer b/w the clathrin cage and the membrane
AP complexes bind transmembrane cargo receptors (eg. LDL receptor), which bind soluble cargo inside the lumen
so AP complexes select which cargo gets packaged
once adaptors bind receptors, clathrin triskelia assemble on them; clathrin only binds via adaptor proteins (can’t bind membrane directly)
there are several types of adaptor proteins, each specitfic for a subset of receptors
Arf is a small GTPase that helps recruit AP complexes
belongs to family of small monomeric GTPase
as more clathrin triskelia assemble, the membrane begins to curve
Clathrin Vesicle Formation: Step 1 Figure
Different AP Complexes
AP1: TGN to endosomes
AP2: endocytosis from the plasma membrane (bud inward for outside material)
AP3: lysosome-related organelles
AP4: specialized pathways
Clathrin Vesicle Formation: Step 2 Fission and Uncoating
dynamin forms a ring around the neck of a budding vesicle
dynamin is a soluble cystolic protein that contains a GTPase domain, which regulates the rate of pinching off
dynamin uses GTP→GDP to change shape to tighten the ring
as dynamin tightens, the inner leaflets of the pinched membrane come close enough to fuse, separating the vesicle from the membrane
dynamin recruits other proteins at the budding vesicle to help bend the patch of membrane (distort the bilayer structure, change lipid composition)
once released from the membrane, the vesicle rapidly loses its clathrin coat under the action of ARF GTPase
the clathrin monomers and AP proteins are recycled
Clathrin Vesicle Formation: Step 2 Fission and Uncoating FIGURE

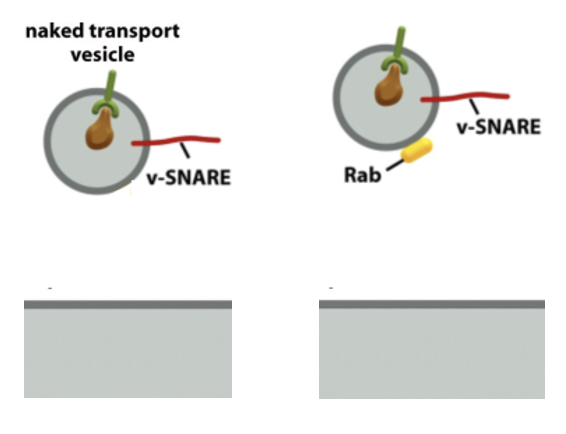
Clathrin Vesicle Formation: Step 3 Recruitment of Rab GTPase
Rab GTPases are membrane-bound proteins
Rab proteins bind to Rab effectors on the target membrane, pulling the vesicle close for fusion
some Rab effectors are tethering proteins that can extend 200nm above the membrane surface
PIP (inositol lipids) are important for the selective distribution
Clathrin Vesicle Formation: Step 3 Recruitment of Rab GTPase FIGURE
Rab Proteins
the largest subfamily of small GTPase (60 members)
the selective distribution of Rab proteins at the surface of the vesicles guides vesicular
PIPs + Rab GEFs Control Rab Binding Specificity
different organelles have distinct PIP compositions; the specific lipid composition helps recruit the right Rab proteins
lipid kinases and phosphatases control PIP levels (located on cystolic face, some integral, some peripheral)
kinases: add phosphate groups
phosphatases: remove phosphate groups
RAB GEFs activate Rab GTPases, then Rab-GTP can bind specifically to membranes with the correct PIP signature
ensures vesicle targeting specificity
PIPs + Rab GEFs Control Rab Binding Specificity
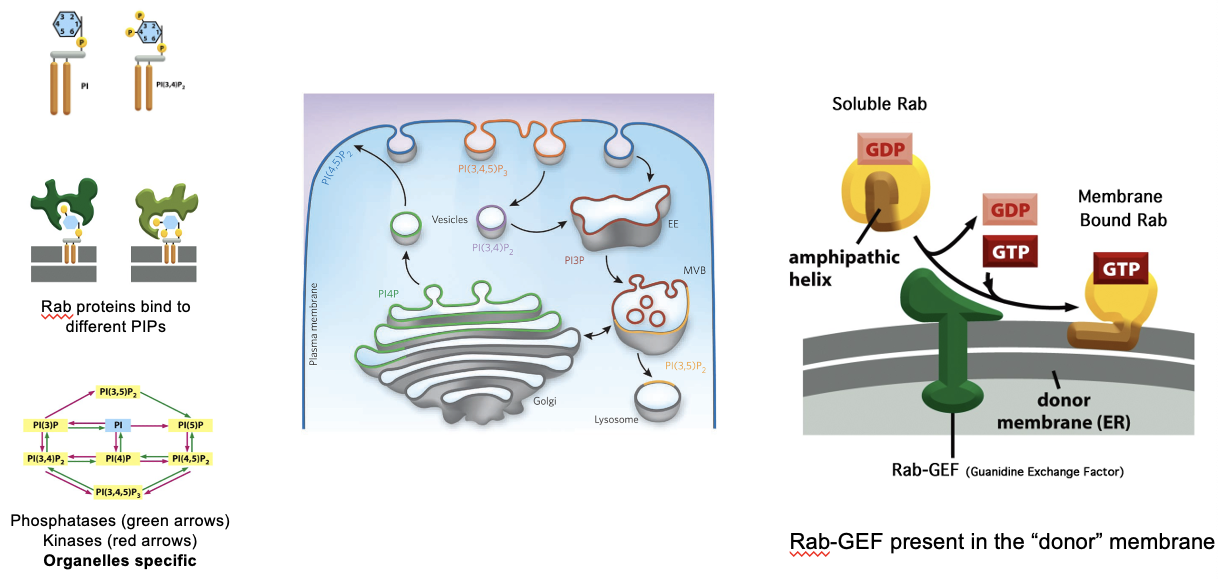
Different PIPs for Different Organelles
early endosome: PI3P
PM: PI(4,5)P2, PI(3,4)P2, PI(3,4,5)P3
late endosomes, lysosomes: PI(3,5)P2
Golgi: PI4P
Forces Applied by SNAREs
SNARE motif: ~60-70 a.a’s with heptad repeats (a repeating pattern of 7 amino acids that allow coiled- coil formation, ⍺-helices wind around each other like the strands of a rope)
zipper model: a v-SNARE (on vesicle) and t-SNAREs (on target membrane) come together
they start pairing from the N-terminal ends (far from the membrane) and zipper toward the C-terminal ends
the zippering process pulls the the two membranes together
membranes naturally repel each other, SNARE zippering overcomes these repulsive forces
when membranes get to ~1nm apart, spontaneous fusion can occur
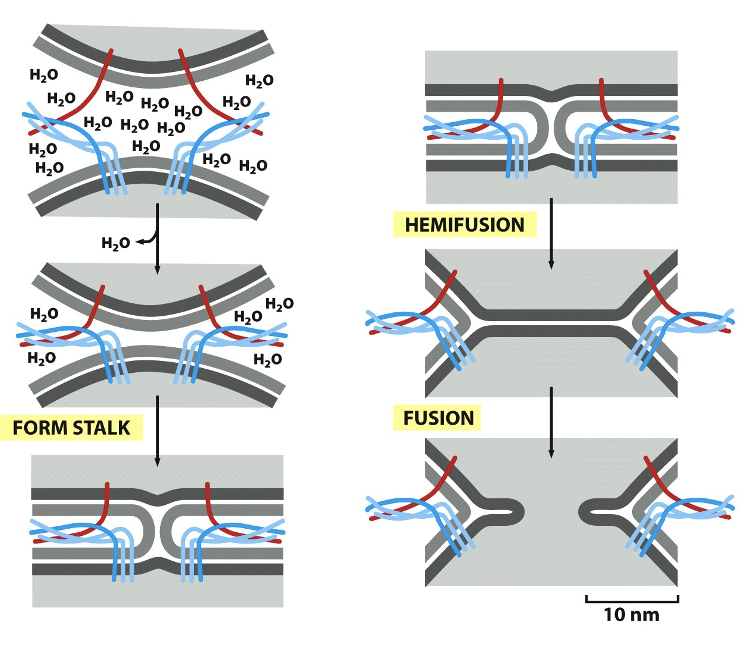
Forces Applied by SNAREs: Energy
SNARE complex formation releases a lot of energy
strong enough to:
displace water b/w membranes
deform the lipid bilayers
form a hemifusion stalk
drive full fusion pore opening
Forces Applied by SNAREs: Figure
Clathrin Vesicle Formation: Step 5 Recycling SNAREs
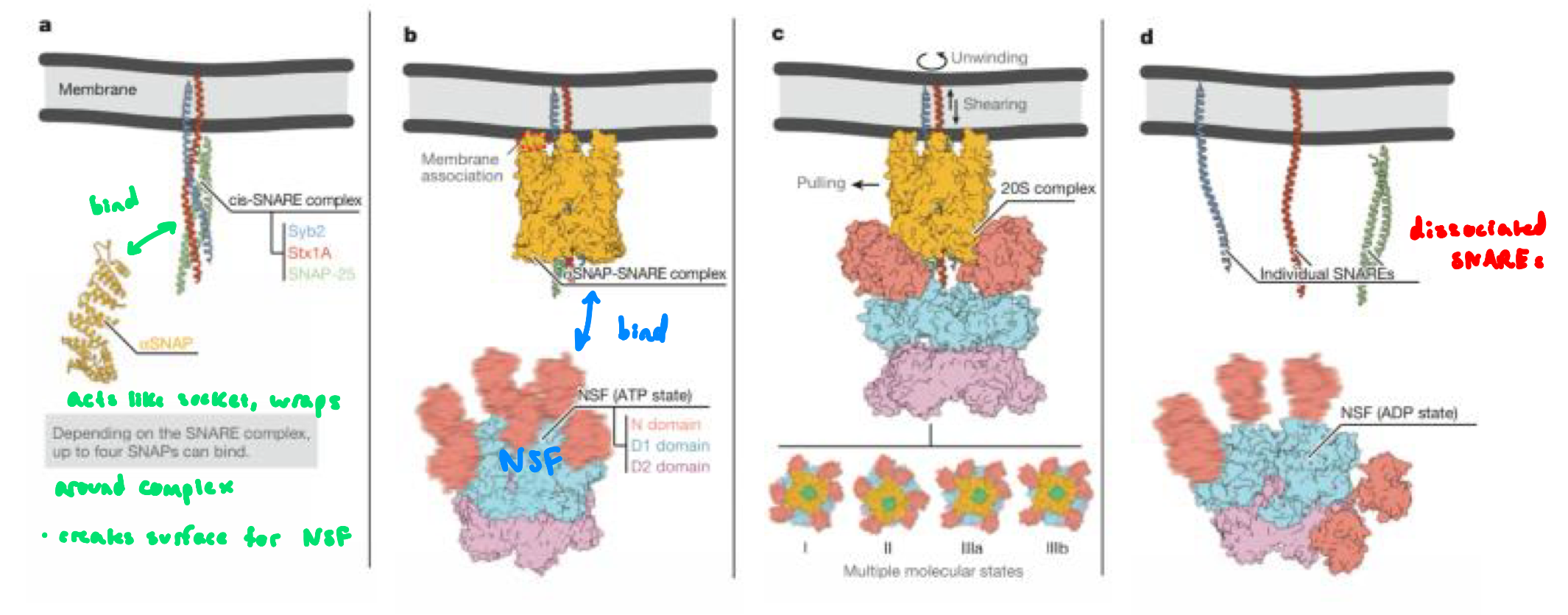
after fusion, the v-SNARE and t-SNAREs are now stuck tgt in. a very stable 4-helix bundle
the SNAREs must be pulled apart so each one can be reused in another round of vesicle fusion
disassembly mediated by NSF (ATPase) and ⍺-SNAP (binds to SNAREs and recruits NSF) proteins
NSF uses energy from ATP hydrolysis to forcibly unwind the SNARE complex
v-SNAREs are then returned to the appropriate membrane and t-SNAREs remain at their membrane
NSF
N-ethylmaleimide-sensitive factor
uses multiple cycles of ATP hydrolysis to pry apart SNAREs
hexamer of identical subunits (termed AAA ATPase) that associates with ⍺-snap (soluble NSF attachment protein)
NEM
N-ethylmaleimide
blocks NSF function
reacts with free SH group on cysteine residues
SNAREs Disassembly Mechanism: “Socket and Wrench”
⍺-SNAP is the socket that attaches onto the assembled SNARE complex
wraps around the complex and creates surface for NSF
NSF is the wrench that clamps onto the socket and turns (using ATP)
together, they produce mechanical force that unwinds the extremely stable helix bundle (V-SNARE and T-SNARE after membrane fusion)
SNAREs Disassembly Mechanism: “Socket and Wrench” FIGURE
Botulinic Toxin from Clostridium botulinum
botulinum toxin and tetanus toxin are neurotoxins produced by Clotridium species that cleave SNARE proteins
these toxins block synaptic transmissions
if SNAREs are cut → synaptic vesicles cannot fuse → no neurotransmitter release
botulinum toxin blocks acetylcholine release at neuromuscular junctions → muscles cannot contract
botox destroys SNAREs preventing muscular contraction
tetanus toxin blocks inhibitory neurons in the central neural system → constant muscle contraction
LD50 ~ 1ng/kg (extremely potent), making it deadly
SNARE Complex
formed by four ⍺-helices contributed by synaptobrevin, syntaxin and SNAP-25
these provide the mechanical force to bring membranes together
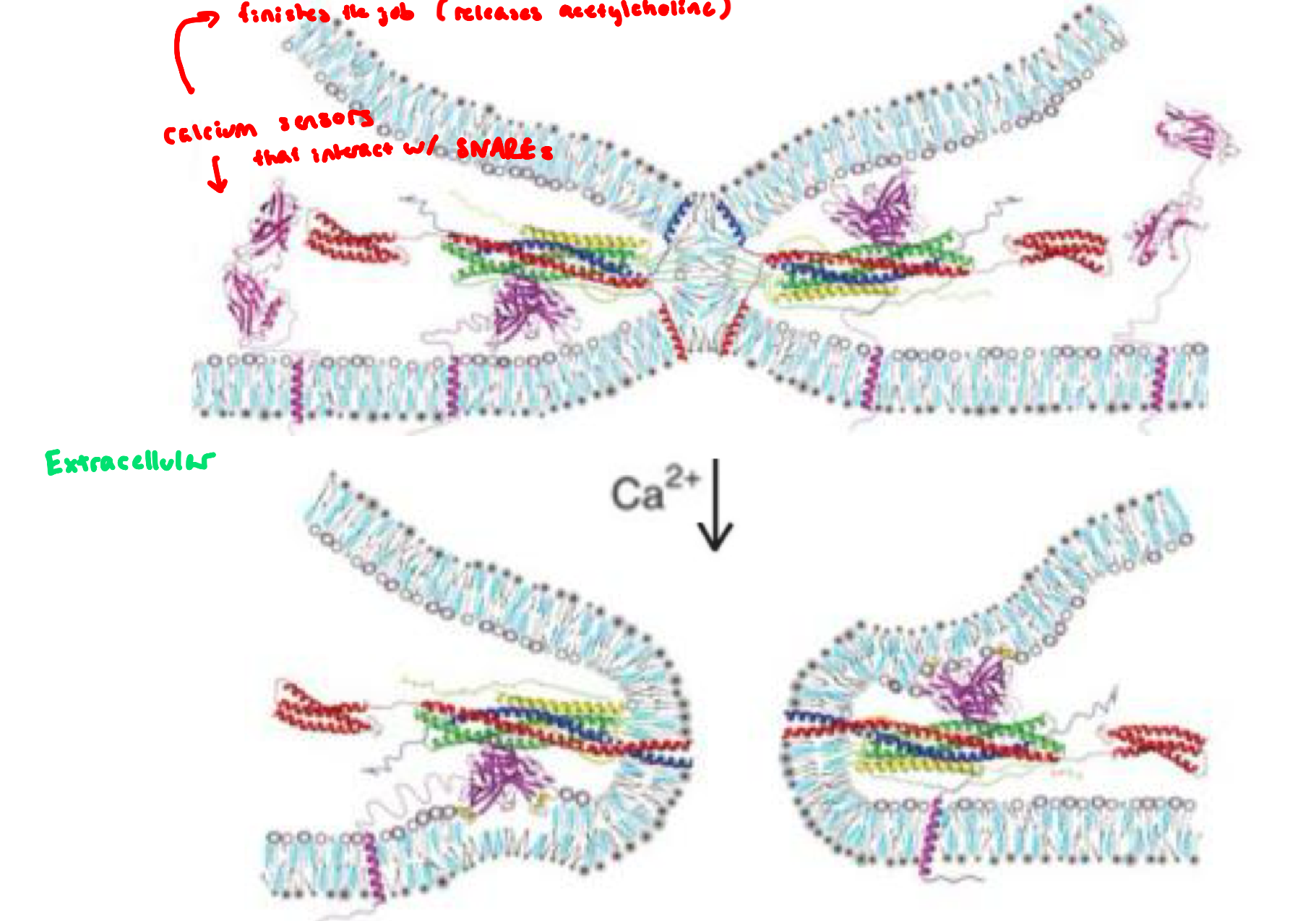
Calcium Binding to Synaptotagmin Triggers Fusion
synaptotagomin: a Ca2+ sensor embedded in the synaptic vesicle and controls whether fusion goes forward
before Ca2+ arrives, synaptotagmin acts as a fusion clamp,
preventing premature fusion when vesicles are docked and primed, and the SNARE complex is partially zippered
synaptotagmin binding to the SNARE complex causes the fusion clamp to tighten further and creates additional disturbance in the bilayer
when Ca2+ enters the neuron, Ca2+ binds synaptotagmin
Ca²⁺-bound synaptotagmin releases the “clamp,” accelerates SNARE zippering, and triggers immediate vesicle fusion → neurotransmitter release
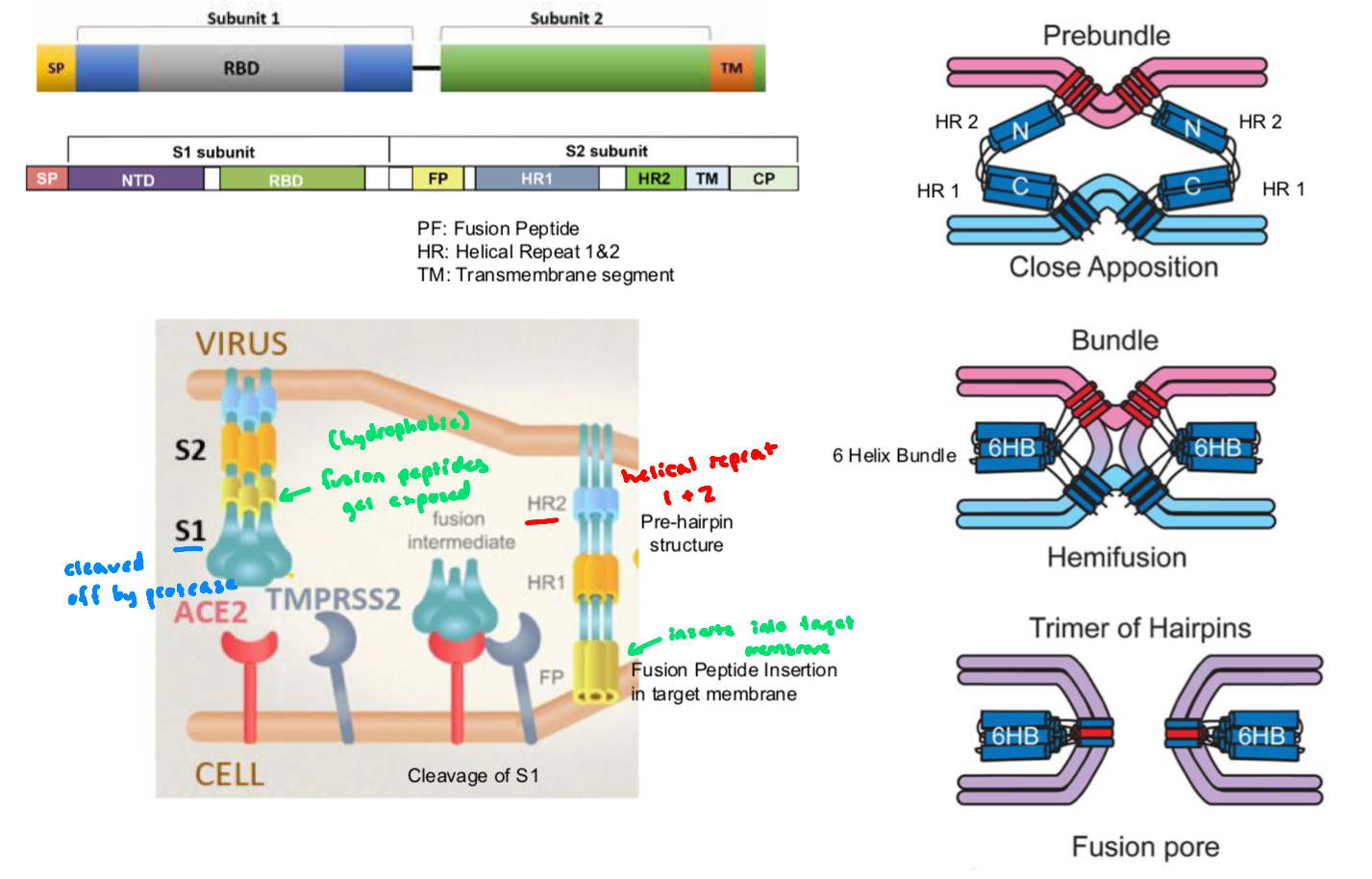
How the Spike Protein Works: SARS-CoV
mediates viral entry and consists of 2 subunits
S1 domain: forms the outer portion of the ectodomain, contains the receptor-binding domain
responsible for recognition and binding to the host cell receptor (Ace-2)
S2 domain: responsible for fusion,
contains the putative fusion peptide (inserts into the host membrane after activation)
and the heptad repeat HR1 and HR2, forming a six-helix bundle that pulls viral and host membranes together
activation by host protease TMPRSS2 (transmembrane serine protease), cleaves the spike protein at the S1/S2 site, priming the spike protein and exposing the fusion peptide → S2 undergoes conformational change that drives membrane fusion
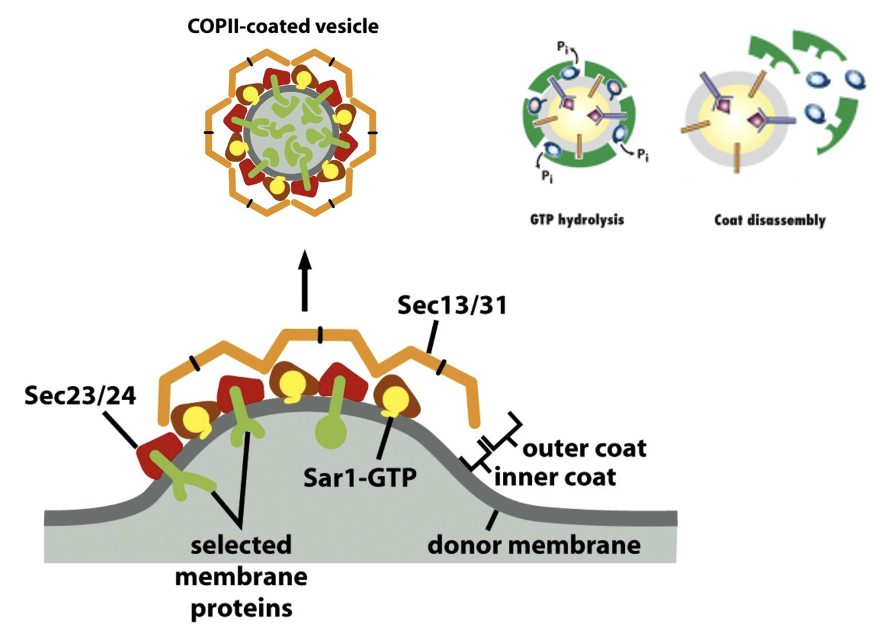
COPII Vesicles
anterograde (ER→Golgi)
carry newly synthesized proteins toward the Golgi
COPII vesicle formation looks similar to clathrin coat assembly/disassembly
COPII Vesicle Formation
SAR1-GTP inserts into the membrane and recruits the inner coat proteins Sec23/24
clathrin system: ARF1-GTP
cargo selection: Sec24 recognizes cargo sorting signals in transmembrane cargo receptors
clathrin: AP (adaptor proteins)
coat assembly + bud formation: Sec23/Sec24 inner coat recruits Sec13/Sec31 outer coat → cage forms
clathrin: AP proteins recruit triskelia
fission is less dependent on dynamin-like protein but still involves curvature forces driven by the coat
uncoating: Sar1 hydrolyzes GTP → coat disassembles after vesicle buds
clathrin: ARF-GTP hydrolysis
COPII Vesicles FIGURE
Fusion of COPII vesicles to form the ERGIC (ER-Golgi Intermediate Compartment)
what happens after COPII uncoat
uses the same Rab+SNARE machinery as clathrin
COPII vesicles bud from the ER, but do not fuse directly with the Golgi, instead they first fuse with each other to form a collection of tubules and vesicles near the ER
the ER-Golgi Intermediate Compartment (ERGIC)
ERGIC moves toward the Golgi, then fuses with cis-Golgi

COPI Vesicles
retrograde (Golgi → ER, or earlier Golgi)
return escaped ER-resident proteins (eg. BiP or ER-resident proteins)
recycle Golgi enzymes, leaving compartments organized
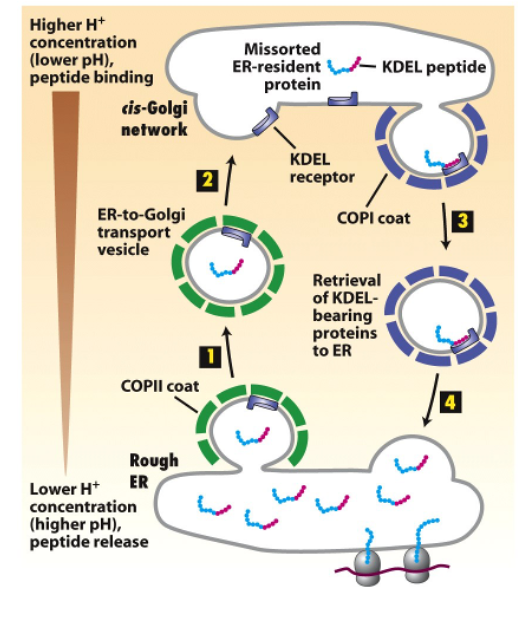
Retrieval of ER-resident proteins
resident ER membrane proteins contain signals that bind directly to COPI coats (KKXX at C-terminus)
soluble ER resident proteins (e.g BiP) contain KDEL signal (Lys-Asp-Glu-Leu)
they bind to KDEL receptor
the affinity of the KDEL receptor depends on the environment (pH sensitive)
the KDEL receptors sits in the Golgi membrane and bind in acidic Golgi pH, releases them in neutral ER pH
this prevents the KDEL sequence from interacting with the KDEL receptor in the ER

pH Controls the Affinity of KDEL Signal Figure
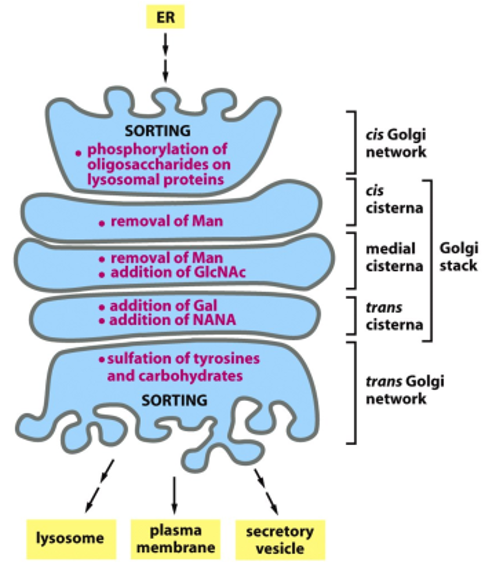
Golgi Apparatus
allows for post-translational modifications (glycosylation, phosophorylation, sulfation)
each Golgi stack has 2 distinct faces: a cis face (entry) and a trans face (exit)
each stack (cisterna) contains a characteristic set of processing enzymes
the Golgi apparatus generates heterogenous oligosaccharide structures, and complex oligosaccharides are added to proteins in the Golgi
the human genome encodes many different Golgi glycosyl transferases
the Golgi resident proteins (glycosidases and glycosyl tranferases) are all membrane-bound
this way the retrieval is facilitated via the COPI mechanism
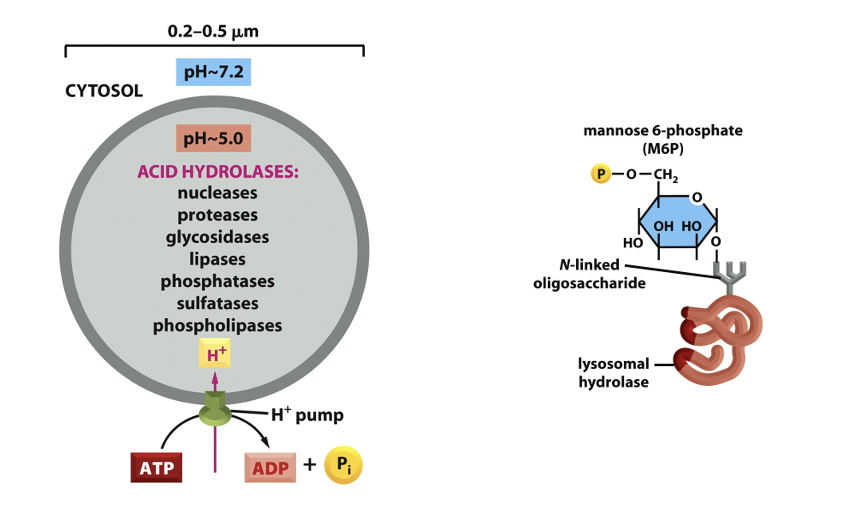
The Lysosome
membrane-bound organelle in the cytoplasm of eukaryotic cells containing degradative, hydrolytic enzymes
the lysosomes membrane is highly glycosylated (needs to protected from the proteases and lipases)
sites of intracellular digestion, serve to digest macromolecules
about 40 types: proteases, nucleases, glycosidases, lipases, phosphatases, sulfatases
proteins are synthesized in inactive form (proenzyme) and require an acidic environment for activation
vacuole H+ ATPase uses the energy of ATP to pump H+ into the lysosome
the H+ gradient provides energy for the transport of metabolites out of the organelle
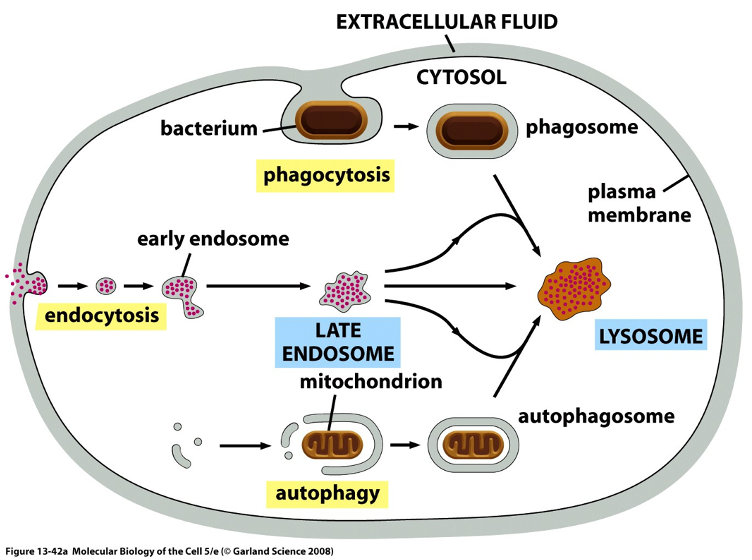
More Lysosome Figures
Delivery of Material to Lysosome: Intracellular Traffic
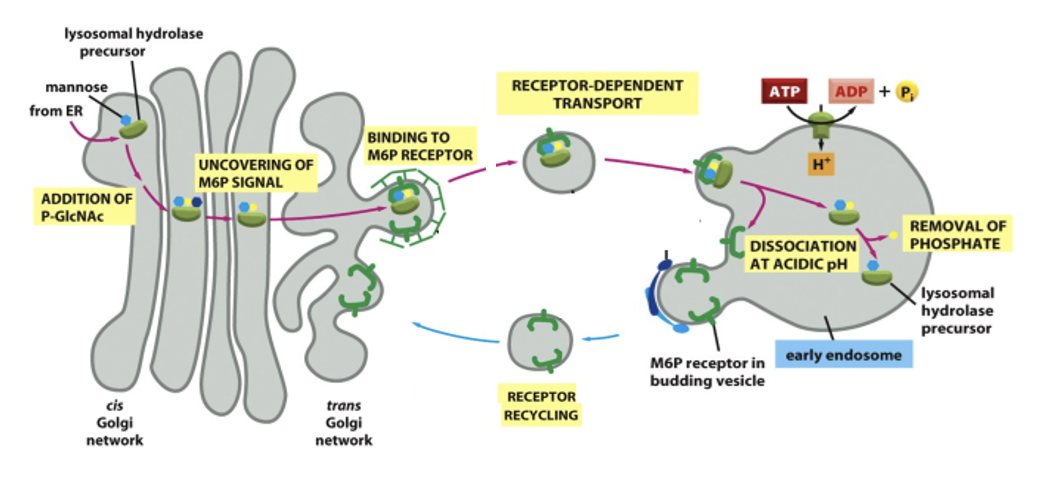
Delivery of enzymes from the Golgi
the enzymes are made in the ER, processed in the Golgi and then tagged with mannose-6-phosphate (M6P)
M6P receptors in the trans-Golgi network package them into vesicles heading toward endosomes → lysosomes
Delivery of cargo from endocytosis
macromolecules (eg. LDL, proteins) enter an early endosomes thru endocytosis
as the compartment acidifies, the early endosome becomes a late endosomes then matures into lysosomes
involves increasing acidity, acquisition of lysosomal enzymes from Golgi, recycling of endosomal membrane proteins back to the PM or TGN
Delivery of Material to Lysosome: Autophagy
Cells digest their own organelles or parts of the cytoplasm
damaged organelles (eg. mitochondria every 10 days in liver cells, bulk cytoplasm (during starvation)
a double-membrane structure forms around the material to form autophagosome, which fuses with lysosome/ late endosome
contents are degraded and the breakdown products (a.a’s, lipids, sugars) are reused
Metabolites derived from the digestion of the captured material help the cell survive
Delivery of Material to Lysosome: Phagocytosis
for large particles and microorganisms
primarily occurs in macrophages and neutrophils
cell engulfs large particles/objects to form a phagosome
phagosome fuses with a lysosome → particle is digested
The M6P Signal binds to M6P Receptor in the Golgi
the M6P-tagged hydrolase binds to the receptor in the Golgi → packed in clathrin-coated vesicles for transport to endosomes/lysosomes
the vesicle fuses with the lysosome and the low pH in the lysosome cause the hydrolase to release from the receptor
an acid phosphatase destroys M6P, prevents the enzyme from rebinding the receptor, keeping it in the lysosome
M6P receptors are retrieved into coated transport vesicles
Lysosomal Storage Disease
autosomal recessive disorder, no cure currently
the enzyme GIcNAc-phosphotransferase (located in the cis-Golgi network) is defective or absent
this enzyme is required to add the M6P tag onto lysosomal hydrolases
if M6P is not added, lysosomal hydrolases cannot bind to M6P receptors in the TGN → do not enter vesicles destined for lysosomes
instead follow default secretion pathway and are secreted outside the cell
undigested substrates accumulate in lysosomes, resulting in inclusion cells (composed of carbohydrates, lipids, other materials that should be degraded)
Lysosomal Storage Disease: Symptoms
Developmental delay
Skeletal abnormalities
Organ dysfunction
Shortened lifespan