BIOTECHNOLOGY & DNA TECHNOLOGY
1/37
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
38 Terms
Define biotechnology and recombinant DNA (rDNA) technology.
Biotechnology: Use of microorganisms, cells, or cell components to make products (e.g., food, enzymes, antibiotics).
rDNA technology: Insertion or modification of genes to produce desired proteins—core of genetic engineering.
What are the major steps in a genetic-modification procedure?
Isolate or obtain a vector.
Obtain or amplify the gene of interest.
Insert the gene into the vector.
Introduce the recombinant vector into a host cell.
Select clones carrying the recombinant DNA.
Use or analyze the resulting gene product.
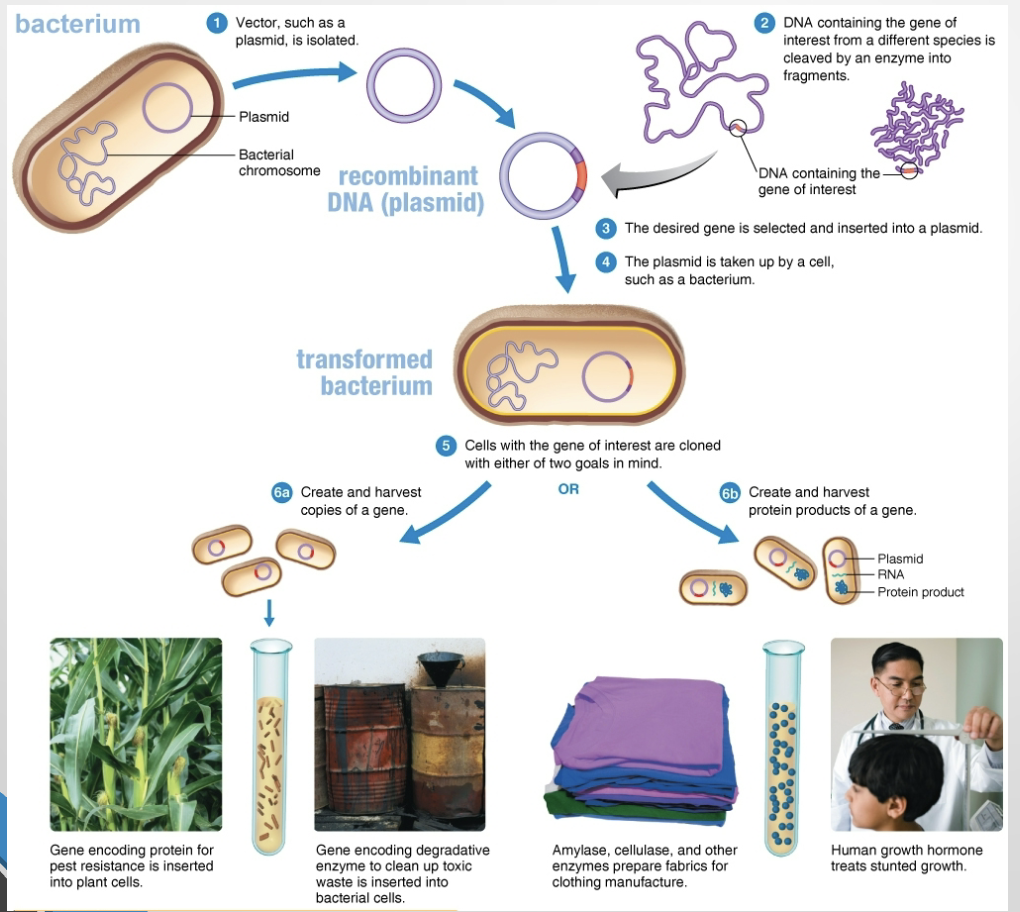
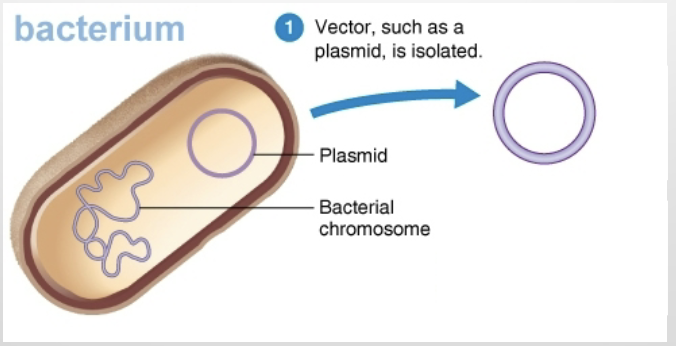
What is a vector?
A self-replicating DNA molecule that transports foreign DNA into a cell (plasmids and some viruses are common vectors).
What is a clone in biotechnology?
A population of genetically identical cells derived from one ancestral cell; each contains the same recombinant vector.
What are shuttle vectors?
Plasmids engineered to replicate in multiple species, allowing movement of cloned sequences between different organisms.
List the four key components of a typical vector.
ORI: Origin of replication.
Multiple cloning site (MCS): Region cut by various restriction enzymes.
Selectable marker: Antibiotic-resistance gene for screening transformants.
Promoter / regulatory elements: Control when and how the inserted gene is expressed.

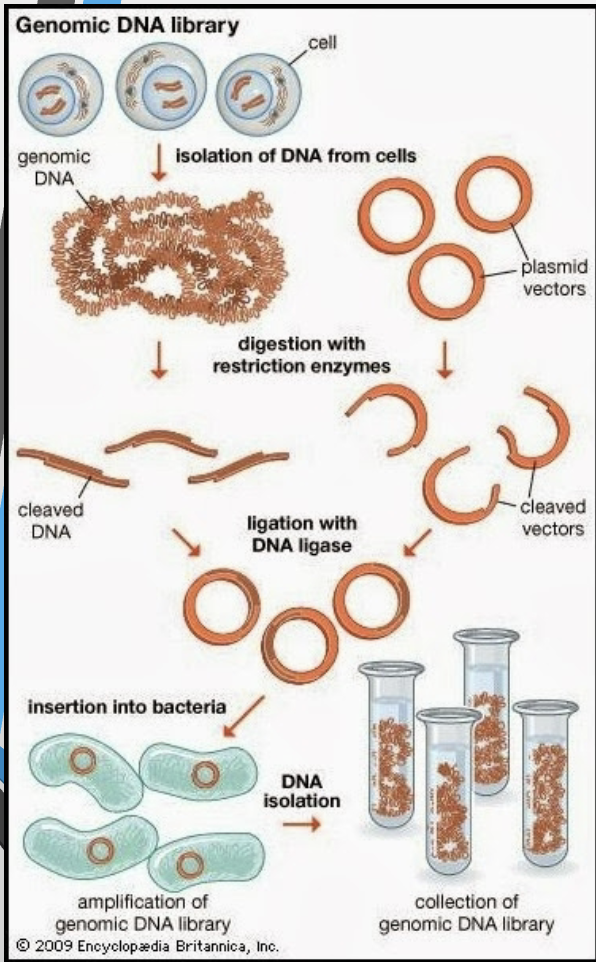
What is a genomic library?
A collection of clones containing fragments of an organism’s DNA inserted into vectors; together they represent the whole genome.
Why is complementary DNA (cDNA) used instead of genomic DNA for cloning eukaryotic genes?
cDNA is synthesized from mature mRNA and therefore lacks introns, allowing expression in prokaryotic hosts that cannot splice introns.
Outline the steps to make cDNA from mRNA.
Isolate mRNA.
Use reverse transcriptase to build a DNA strand.
Degrade mRNA.
Add DNA polymerase to synthesize the second strand.
→ Double-stranded cDNA ready for cloning.
What is synthetic DNA and when is it used?
DNA chemically built base by base using a synthesis machine when the sequence is known; used for short genes or designed constructs.
What is PCR and why is it important?
An in-vitro process that exponentially amplifies a specific DNA segment for cloning, diagnostics, or pathogen detection. Makes millions of copies
What are the five basic components of a PCR reaction?
Template DNA → what you’re copying.
Primers → short synthetic DNA sequences that mark the start and end.
Taq DNA polymerase → heat-stable enzyme from Thermus aquaticus.
dNTPs → the nucleotide building blocks (A, T, G, C).
Buffer with Mg²⁺ → keeps the enzyme active and pH stable.
Describe the three core steps of one PCR cycle.
Denaturation (94 °C): DNA strands separate.
Annealing (50–60 °C): Primers bind to target sites.
Extension (72 °C): Taq polymerase adds nucleotides.
Why is Taq polymerase used in PCR?
It is thermostable, isolated from Thermus aquaticus, and remains active at the high temperatures needed for DNA denaturation.
What are two important PCR variants and their uses?
Site-directed mutagenesis: Creates intentional base substitutions or deletions.
RT-qPCR: Converts mRNA to cDNA and quantifies amplification to measure gene expression.
What are restriction enzymes and their natural function?
Bacterial enzymes that cut DNA at specific recognition sequences to destroy invading phage DNA; host DNA is protected by methylation.
How are restriction enzymes used in the laboratory?
They cut DNA precisely at recognition sites to generate fragments that can be joined into vectors, enabling recombinant DNA construction.
What are sticky ends vs. blunt ends?
Sticky ends = single-stranded overhangs that pair with complementary sequences.
Blunt ends = straight cuts without overhangs.
Why are sticky ends preferred in cloning?
They ensure directional insertion and more efficient ligation.
Summarize the steps showing a restriction enzyme’s role in making rDNA.
Cut plasmid and target gene with the same restriction enzyme.
Complementary sticky ends anneal.
DNA ligase joins fragments.
The recombinant plasmid is introduced into a microorganism for propagation.
What is transformation in biotechnology?
Uptake of recombinant plasmid DNA by a host cell from its environment.
How is chemical transformation achieved?
Cells are treated with CaCl₂ to neutralize membrane charge and become competent, allowing DNA entry.
What is electroporation and why is it used?
Electric pulses create transient pores in cell membranes to allow DNA uptake—especially effective after removing cell walls (protoplasts).
Describe protoplast fusion.
Two cells with enzymatically removed walls fuse, allowing their genetic material to recombine; common in plant or algal engineering.
What are gene guns and microinjection used for?
Physical DNA delivery:
Gene gun: Shoots DNA-coated metal particles into cells (plants).
Microinjection: Directly injects DNA into animal nuclei (creates transgenic animals).
Why must selection follow transformation?
Because only a fraction of cells take up plasmids, and not all plasmids contain the desired insert.
How does antibiotic selection work?
Vectors carry a resistance gene (e.g., ampᴿ)—only transformed cells survive on antibiotic-containing medium.
What is artificial selection in this context?
Choosing and propagating cells that exhibit desirable traits, such as growth on selective media indicating successful plasmid uptake.
What is the goal of blue-white screening?
To distinguish colonies with recombinant plasmids from those with empty vectors using color change on X-gal media.
Which plasmid features enable this test?
ampᴿ (ampicillin resistance) and the lacZ gene encoding β-galactosidase.
Explain the principle behind the color reaction.
β-Galactosidase hydrolyzes X-gal to a blue compound; disruption of lacZ by an insert prevents enzyme production → white colonies.
What does a blue colony indicate?
Bacterium carries plasmid without the DNA insert; lacZ intact, enzyme active.
What does a white colony indicate?
The insert disrupted lacZ → no β-galactosidase → no X-gal breakdown → white color; successful recombinant candidate.
Why must ampicillin be included in the agar plate?
To ensure that only cells containing the plasmid grow; non-plasmid cells die.
Summarize the step-by-step process of blue-white screening.
Digest plasmid + target DNA with same restriction enzyme.
Ligate, transform E. coli.
Plate on medium with ampicillin + X-gal.
Blue = empty vector; white = recombinant plasmid.
What molecule acts as the indicator substrate in this assay?
X-gal, a colorless analog of lactose that turns indigo-blue when cleaved by β-galactosidase.
How does the α-complementation system work in this experiment?
The plasmid supplies the α-fragment of lacZ; the host E. coli provides the ω-fragment. Together they make active enzyme unless disrupted by an insert.
What does blue-white screening confirm, and what does it not?
Confirms that a DNA fragment was inserted, but not its sequence accuracy or orientation—those require PCR or sequencing verification.