36 - DNA Transcription
1/50
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
51 Terms
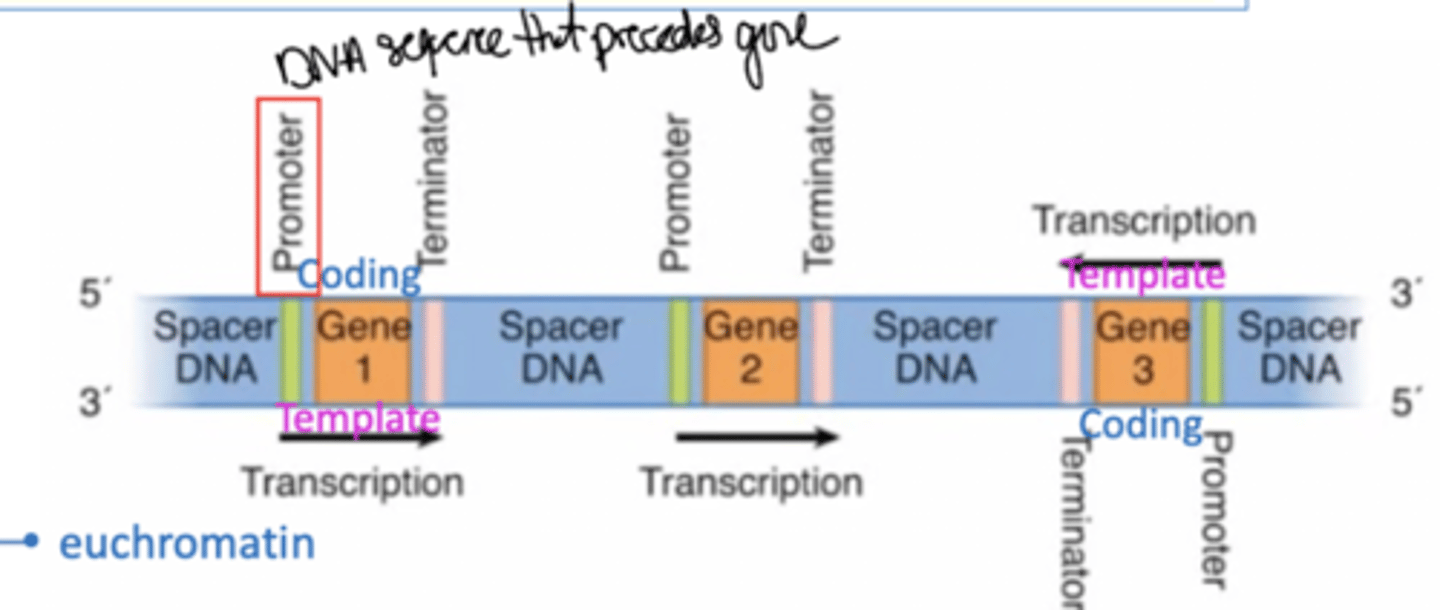
Describe the components of a gene.
What are the three main types of RNA?
tRNA: carries amino acids to translation machinery; very stable
rRNA: makes up much of the ribosome; very stable and makes up the majority of cellular RNA
mRNA: encodes message from DNA to ribosomes; they are rapidly degraded by nucleases
What are the template and coding strands of a gene?
The template strand is used in trancription and is complementary to the RNA. The coding strand is NOT used in transcription and is identical to the mRNA (exceptions being the Ts and Us); easy way to remember is that coding strand "codes" for proteins.
What directions does RNA polymerase operate in?
RNA pol reads the DNA from the 3' to 5' direction. while RNA is synthesized in the 5' to 3' direction
What are some characteristics of RNA polymerase?
RNA polymerase catalyzes DNA-directed RNA synthesis - DNA continuously unwound as RNA pol catalyzes a processive elongation of RNA chain.
It catalyzes polymerization in a 5' to 3' direction. It is highly processive and thermodynamically assisted by PPI hydrolysis.
To which portion of a gene does RNA pol bind?
The promoter sequence. not the gene
What are the conformational changes that occur in RNA polymerase? Why does this occur?
RNA polymerase changes conformation at the initiation site during the unwinding of DNA. RNA pol and the promoter complex shift between RPc (closed complex, while the DNA is stll double stranded), and RPo (open complex, in which 18 DNA bp unwind, forming the transcription bubble)
What are consensus sequences?
Consensus sequences are found upstream from the transcription start site and DNA-binding proteins bind to promoter sequences and direct RNA pol to promoter site
How is transcription terminated?
Transcription sequences disassemble at the 3' end of genes at termination sequences (two types: unstable elongation complex, and rho-dependent termination)
What are pause sites?
Regions of a gene where the rate of elongation slows down - termination often occurs here
G-C regions are more difficult to separate than A-T regions and therefore may be pause sites
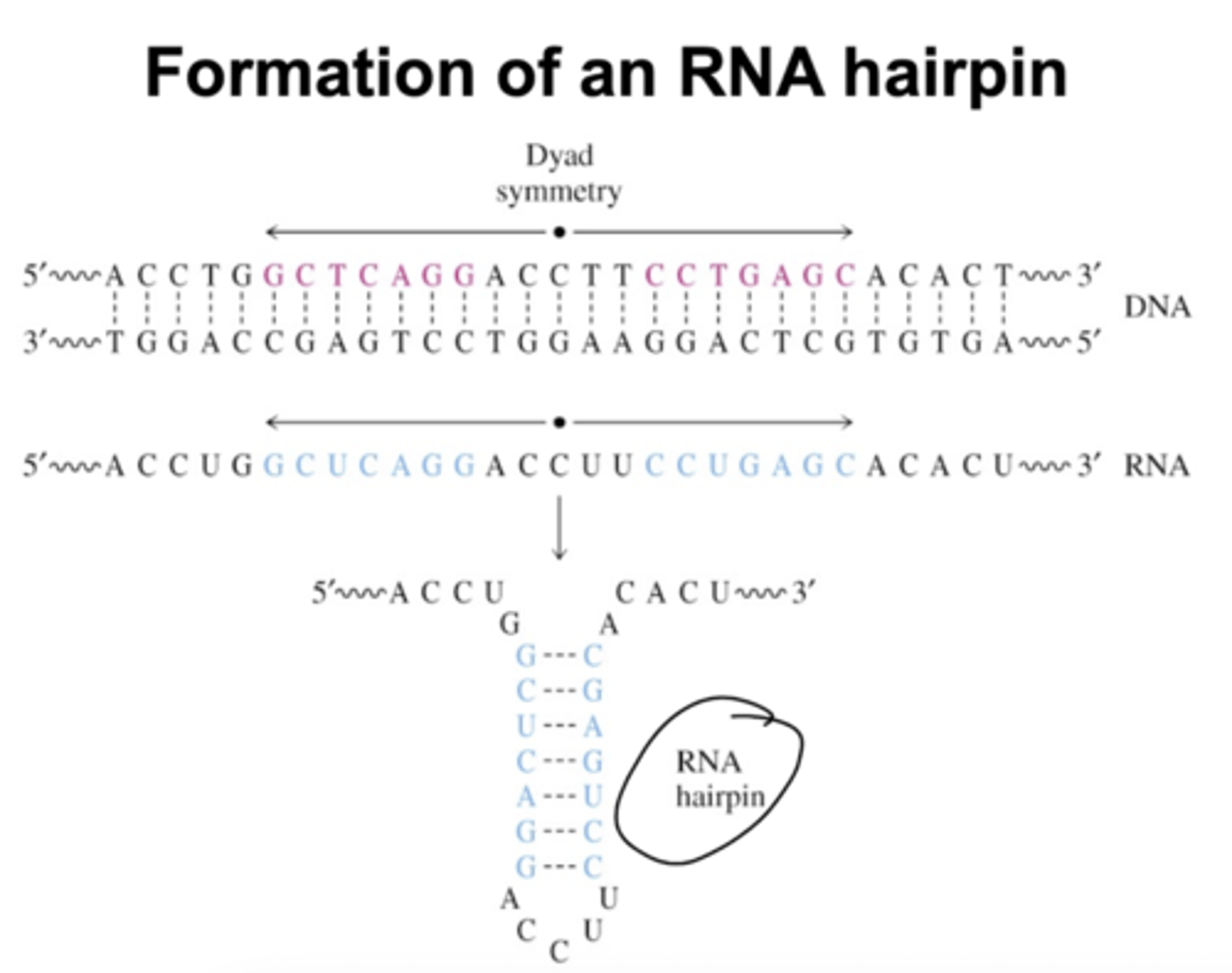
How are RNA hairpins formed?
This is caused by Dyad symmetry in which the DNA is symmetric about a few nucleotides. This leads to RNA forming internal base pairing. This can lead to exaggerated pause sites.
What is constitutive expression?
Refers to the expression of housekeeping genes, which is always active during normal cell function. They have strong promoters and are continuouslt transcribed.
What are repressors?
Regulatory proteins that prevent transcription of a negatively regulated gene - negatively regulated genes can only be transcribed in the absence of a repressor
What are activators?
Regulatory proteins that activate transcription of a positively regulated gene
What are inducers?
Ligands that bind to and inactivate repressors
What are corepressors?
Ligands that bind to and activate repressors
How is transcription initiated?
The transcription complex (including proteins and RNA pol) assembles at an initiation site (DNA promoter region)
What is an operon?
An operon is a transcription unit in which several genes are often co-transcribed in prokaryotes
What is a difference in initiation between eukary and prokaryotic genes?
Eukaryotic genes each have their own promoter, while prokaryotic genes have one promoter for multiple genes
What are some characteristics of the E. Coli promoter?
TATA box (-10 bp upstream from transcription start site)
-35 region (upstream) from start site
-Strong promoters match consensus sequence closely (operons transcribed efficiently)
-Weak promoters match consensus sequence poorly (operons transcribed infrequently)
How does glucose level affect synthesis of lac operon proteins?
Present glucose limits synthesis of the proteins. Low glucose levels increase synthesis of the proteins.
What is the function of operators in operons?
Operators bind repressors in operons to repress the expression of genes in the operon.
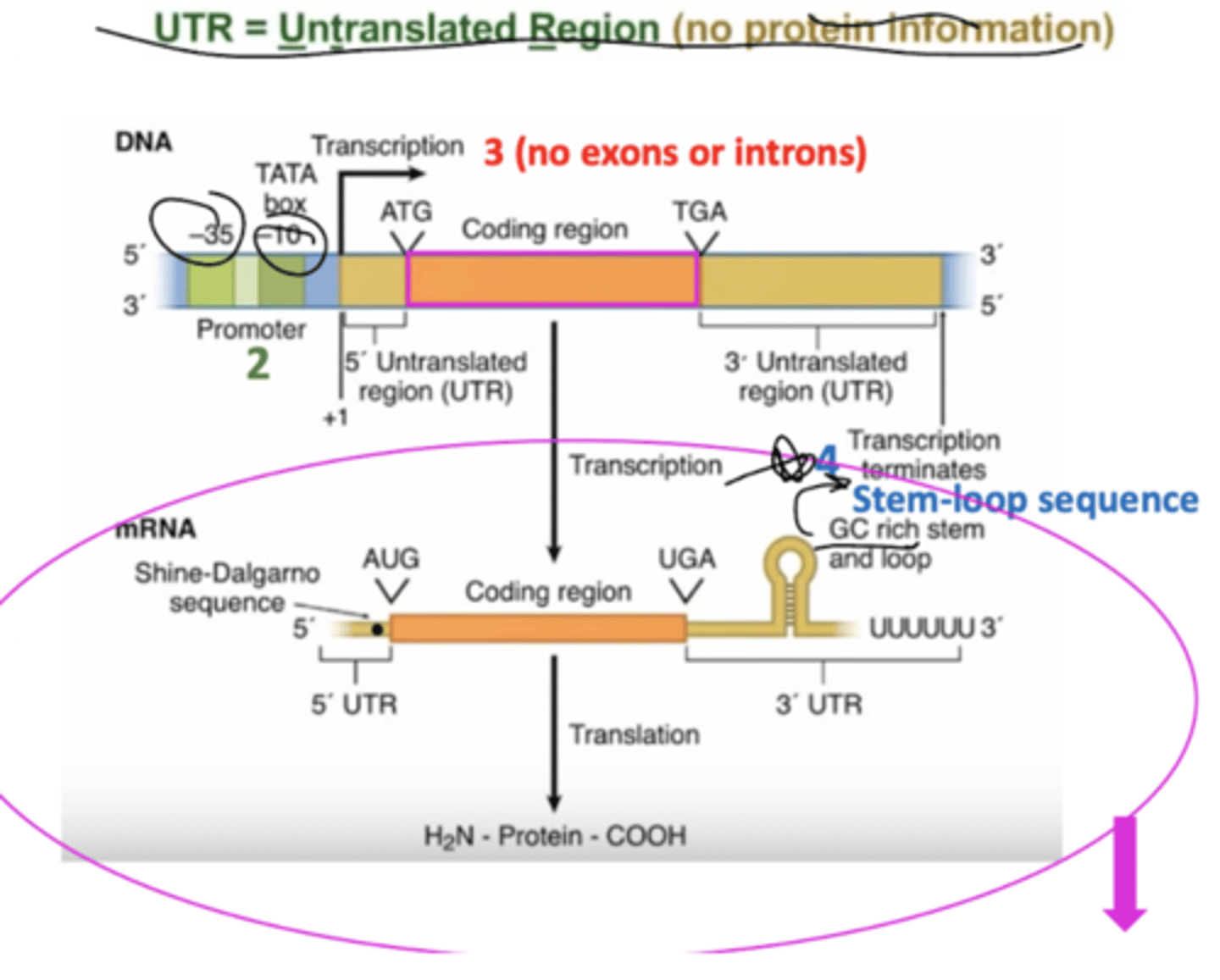
What are some major characteristics of prokaryotic transcription>
An operon is a polygene region consisting of a promoter (2), no exons/introns (3), and a stem-loop sequence (4) that terminates transcription.
What is Fragile X syndrome?
Aka Martin-Bell syndrome. More prevalent in males than females. It results in developmental delays, intellectual disabilities, learning disabilities, anxiety, ADHD, and autism spectrum disorder.
What is the Shine-Dalgarno sequence?
A short conserved nucleotide sequence upstream of the AUG start codon that serves to align the mRNA on the bacterial small ribosomal subunit. It is a part of the 5' UTR of prokaryotic mRNA that is where the ribosome binds.
What is the major characteristic of the genes in an operon?
They are translated independently, creating separate proteins.
What is rifampin?
A drug that inhibits prokaryotic RNA pol -- used in the treatment of tuberculosis and meningitis. This causes red urine.
What are the target of quinolones?
Topoisomerases
How is the 3' end of eukaryotic mRNA modified?
A poly A polymerase adds up to 250 adenylate residues to the 3' end of the mRNA, which is progressively shortened by 3' exonucleases
What is the role of the poly A tail?
It increases the time required for 3' exonucleases to reach the coding region of a strand of mRNA, thus increasing its lifespan
What are introns?
Internal sequences that are removed from the primary RNA transcript
What are exons?
Sequences that are present in the primary transcript and the mature mRNA
What are splice sites?
Junctions of the introns and exons where mRNA precursor is cut and joined
What is the spliceosome? What is it made of?
It is a large RNA-protein complex which catalyzes splicing reactions. It contains 45 proteins and 4 small nuclear (snRNA) molecules.
What are some characteristics of eukaryotic genes?
Each gene is more widely spaced and has its own promoter. There is no operon (1), there is a promoter sequence (2) for each gene, there are intervening introns to allow for diverse coding (3), there are termination sequences (4).
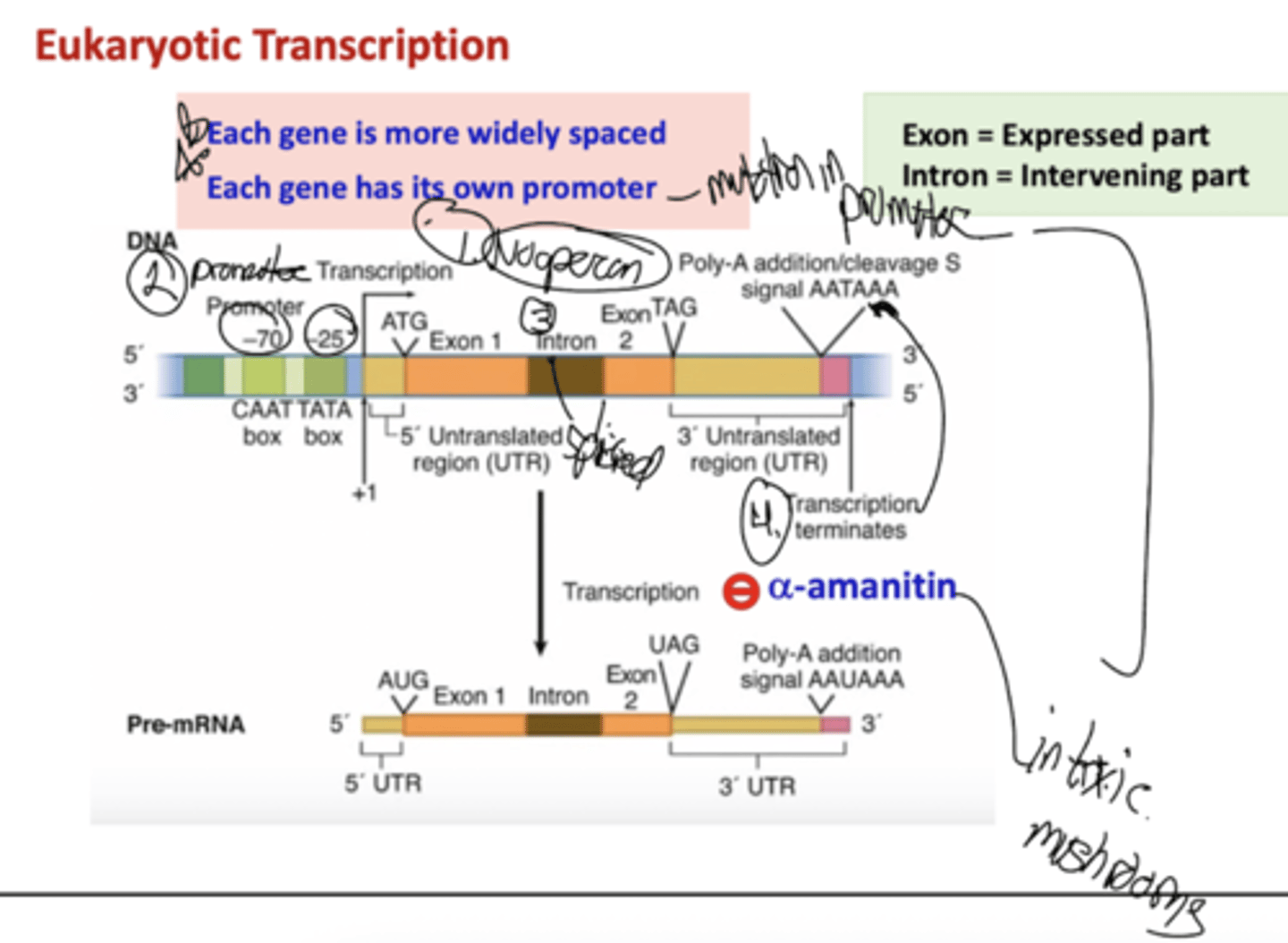
What is a-amantin?
An inhibitor of transcription (RNA pol). It is found in toxic mushrooms
What are some characteristics of eukaryotic transcriptional processing?
Addition of a 7-methylguanylate cap (5.1; this is COTRANSCRIPTIONAL and is where the ribosome binds), post-transcriptional addition of a poly-A tail (5.2; by PolyA polymerase), and splicing by spliceosome (5.3, COTRANSCRIPTIONAL). These occur in the nucleus.
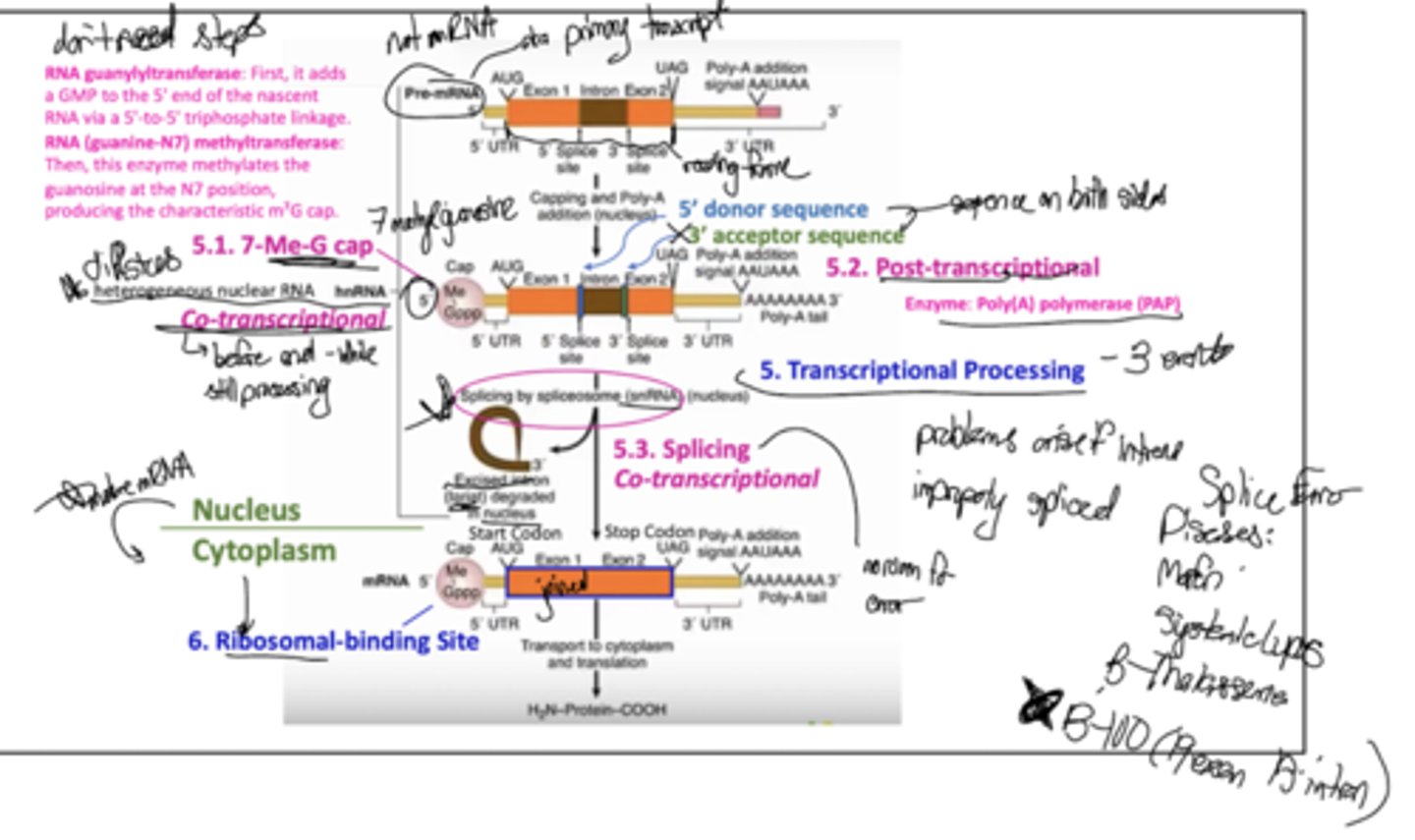
What is the name for an excised intron?
Lariat
What are some splice error diseases?
Marfan, Systematic Lupus, B-thalassemia, and B-100 receptor (because its gene has 19 exons with 18 introns which is wack)
What is alternative splicing? What are some examples?
The removal of different sets of introns and exons in order to transcribe different proteins. This leads to different proteins from one gene, such as membrane and secreted immunoglobins, tropomyosin variants in muscle, and dopamine receptors in the brain
What are some differences between prokaryotic and eukaryotic ribosomes?
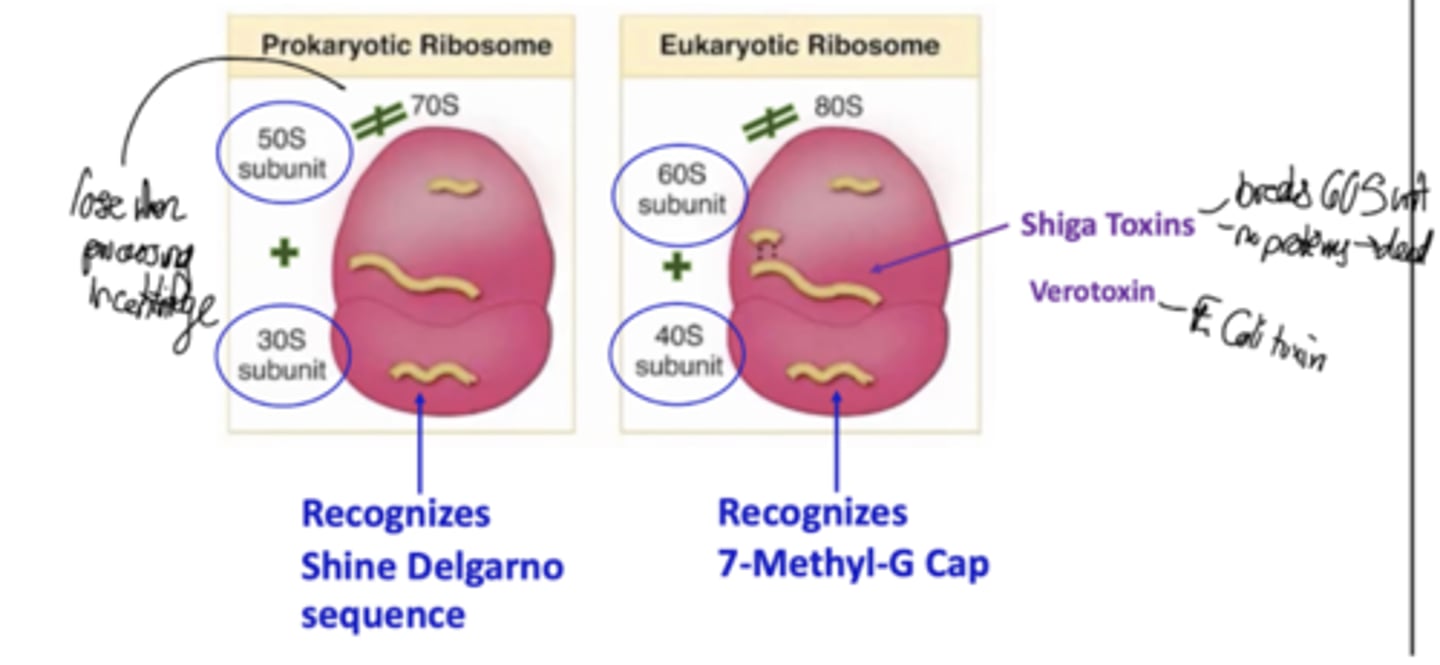
What are some toxins that inhibit the combining of ribosome components?
Shiga toxins on the 60S subunit, and verotoxin on E. Coli 50S
What are some characterisitics of tRNA?
They pick up amino acids and recognize codons with an anticodon. They have a 3' end to which can activated amino acid attaches and a 5' end that is phosphorylated. They have modified bases and an epsilon and a thymine for some reason.
What are the characteristics of DNA mutations?
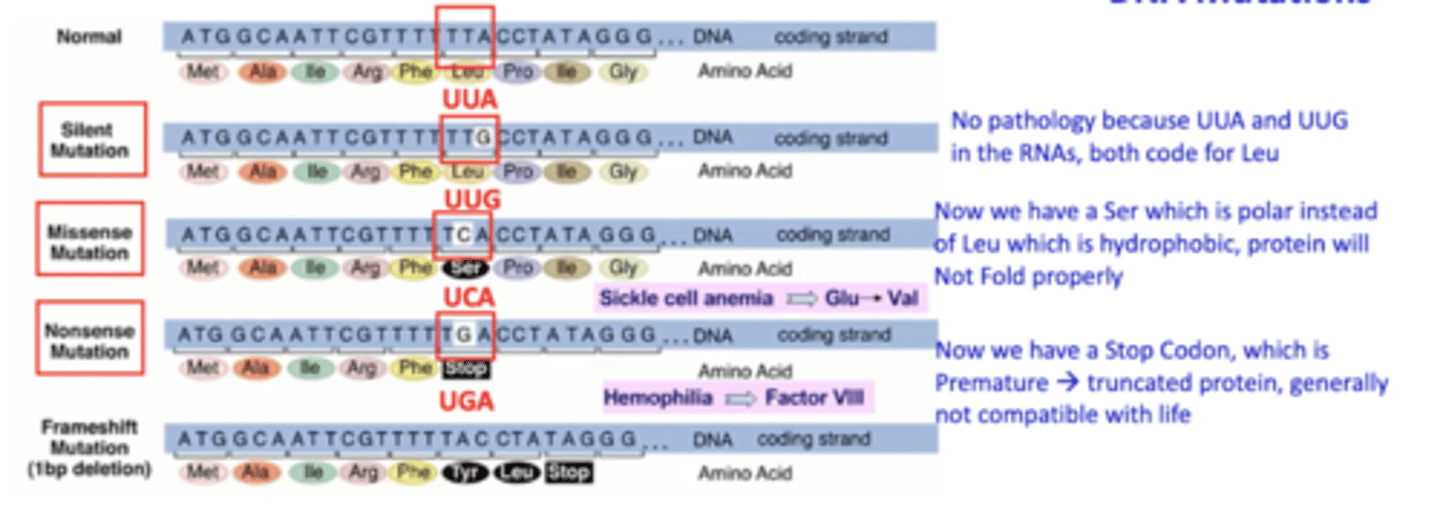
What are the mutations and errors in Duchenne MD?
A deletion causing a frameshift in the dystrophin gene
What are the mutations and errors in cystic fibrosis?
A deletion not causing frameshift in the CFTR gene (delta f508)
What are the mutations and errors in Tay-Sachs?
An insertion causing frameshift in the hexosaminidase A gene
What are the mutations and errors in Huntington's Disease?
Insertion not causing a frameshift in the huntingtin gene
What are the 5 trinucleotide repeat diseases?
Huntington, fragile X, myotinic dystrophy, spinobulbar muscular atrophy, and Freidreich's Ataxia.
Describe the activation of aminoacyl-tRNA. Why must it be activated?
Peptide bonds formation is not energetically favorable on its own, and therefore this activation step ensures the reaction is thermodynamically possible. It takes 2 Pi to form the bond between an amino acid and the 3' end of a tRNA
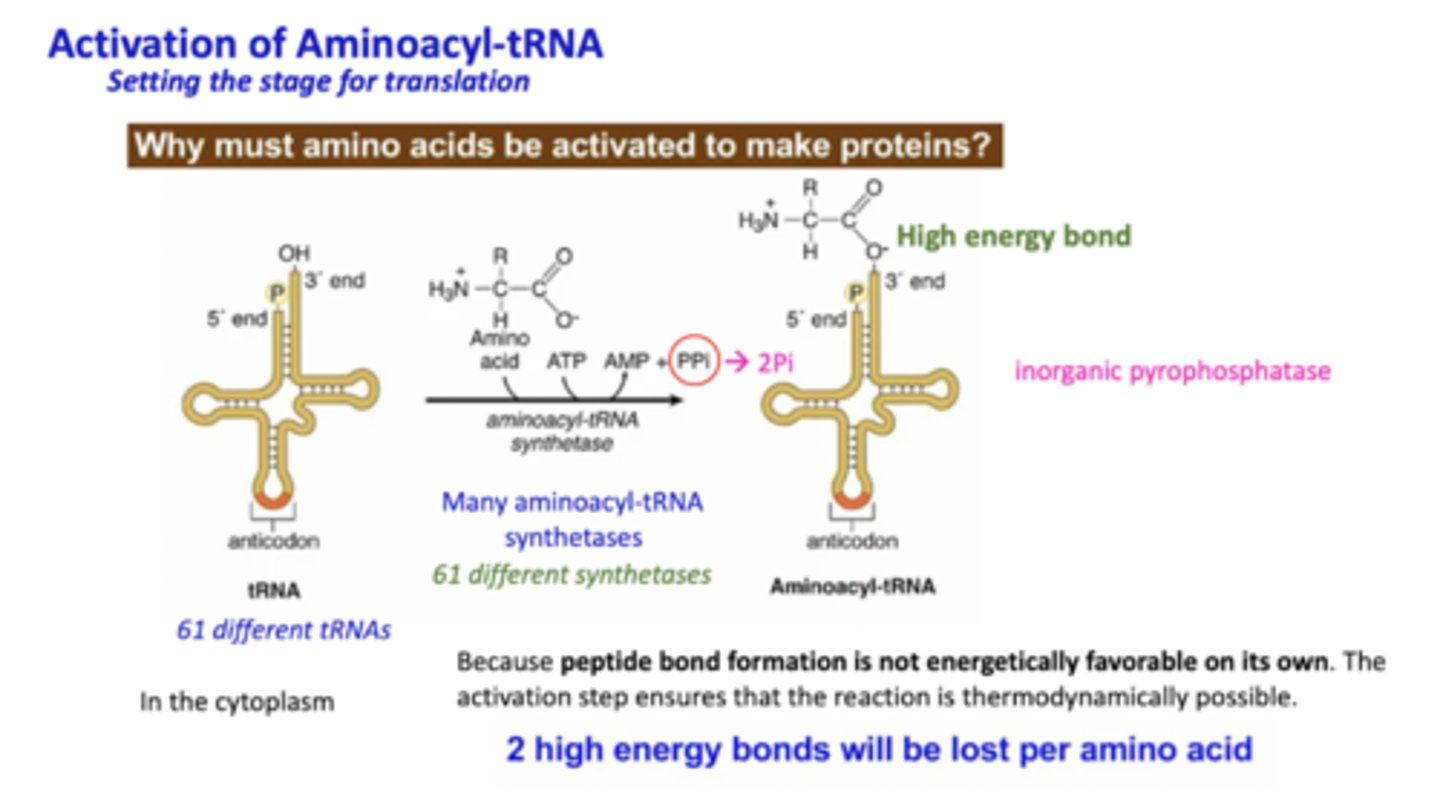
What forms the peptide bond of amino acids?
Peptidyl transferase. It is a ribozyme (not a protein - made of RNA)