D1.1 DNA replication
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
37 Terms
cell cycle
G1:
S phase:
where DNA replication occurs
G2:
Mitosis:
= 2 cells
non-coding regions of DNA
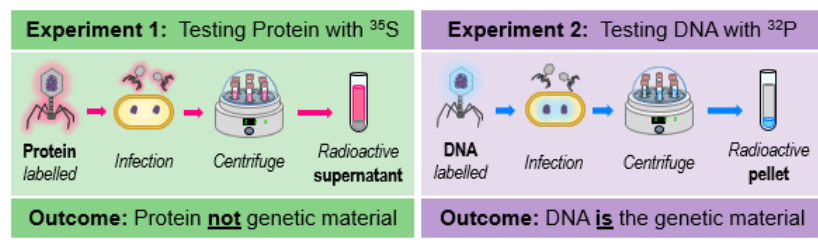
Hershey and Chase experiment?
Hershey & Chase conducted experiments to identify the genetic material of a cell, unknown if DNA or protein was gentic material
used radioactively labeled viruses to infect bacterium (then separated via centrifugation)
• Viruses grown in 355 had radiolabelled proteins in supernatant (not transferred)
• Viruses grown in 32p had radiolabelled DNA in the pellet (transferred to bacteria)
From this it was concluded that DNA is the genetic material
pre-replication protein complex
breaks the first couple hydrogen bonds in the origin of replication to strat forming the rpelication bubble
purpose of DNA replication
needed for cell division(mitosis and meiosis) where cells must replicate their DNA before dividing to ensure each daughter cell has a full set of genetic material(produce diploid or haploid cells).
growth and repair: multicellular organisms grow and repair tissue damage via new cells, each new cell needs it’s own copy of the genome
reproduction(meiosis) to ensure that genetic information is passed down from one generation to the next via sexual/asexual reproduction, continue evolution.
DNA polymerase I
removes the RNA primers on the leading and lagging strands and replaces it with DNA
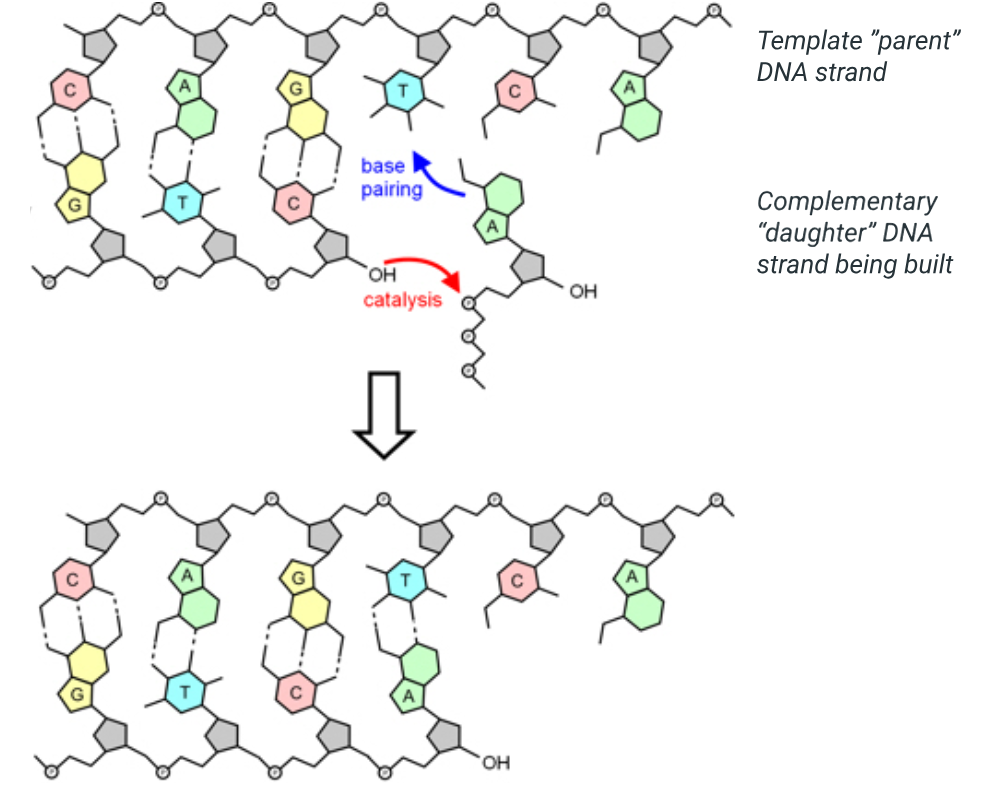
complementary base pairing
for a new base to be added onto a daughter strand it must fit the exposed nitrogenous base ont he aprent strand, as nitrogenous bases pair up based ont he number of available chemical groups where hydrogen bonds can form for each base:
adenine pairs with thymine in DNA or with uracil in RNA, two hydrogen bonds, purines
guanine pairs with cytosine, three hydrogen bonds, pyrimidines
note: if eg. cytosine were to try to pair with adenine which only offers 2 compaitble spots then the electrical charges will not align, so the two repel each other and no stable conenction can be formed
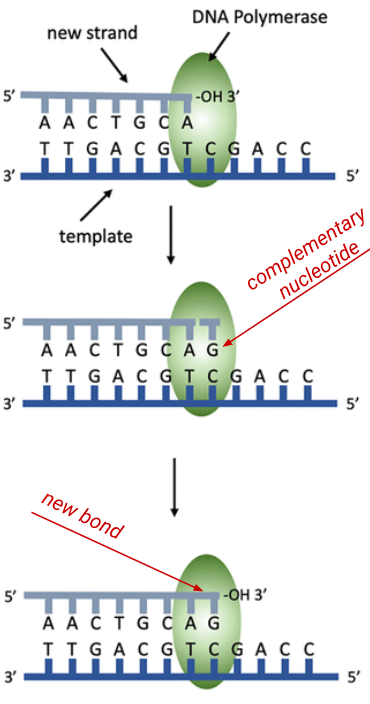
DNA polymerase III can….
only add a new DNA polymerase is chemically specific bc it catalyzes the
condensation reaction that forms the covalent bond between the 5' phosphate group on the "new" incoming nucleotide and the 3' hydroxyl(-OH) group on the sugar of the "last" nucleotide in the chain.
acts as a proofreader of the new daughter strand of DNA bc it can recognise incorrect DNA nucleotides in the daughter strand, reverses direction to remove the incorrect nucleotide from the 3' end of this strand, the correct nucleotide is then inserted, and the polymerase III enzyme continues replication
cell replication
produces two replicas of the parent cell, producing 2 identical daughter cells.
semi-conservative model
use old parental strand to make a new complimentary strand
producing a daughter cell with one old strand and one newly synthesised complementary strand
DNA replication is semi-conservative
directionality: can only add nucleotides onto the 5’ end towards the 3’ end, DNA is bi-directional
helicase
enzyme that breaks the hydrogen bonds between complementary bases in DNA strands to unwind the DNA, splitting the strands up
its movement along the DNA strands is an active process which requires energy fromt he hydrolysis of ATP
how can helicase unzip the strands?
it breaks the hydrogen bonds as it moves along the strands, movement along the DNA strands is an active process which requires energy from the hydrolysis of ATP
leading strand
made continuously, following the fork as it opens, before new DNA nucleotides can be added to the new DNA strand, first an RNA primer must be added to create a binding point for DNA polymerase III
lagging strand
lagging strand is made discontinuously,in short fragments, away from the forkcalled Okazaki fragments
As more template strand is exposed, new fragments are created which are later joined together by DNA ligase to form a continuous complementary DNA strand
The RNA primer only has to be added once on the leading strand but several are needed on the lagging strand to initiate each fragment-
DNA replication
replicate DNA to make new cells
occurs in a semiconservative fashion to make a mixed old and new double-stranded DNA molecule
occurs in a 5’—> 3’ direction
happens bi-directionally from the origin of replication, where the replication forks are(unwinding bothways left/right
DNA polymerase III
DNA polymerase enzymes catalyze the synthesis of the complementary daughter DNA strands.
it moves along the template strand, adding the correct free nucleotide based on complementary base pairing.
The enzyme catalyzes the condensation reaction that creates covalent bonds between the phosphate group of a new nucleotide and the pentose sugar of the previous one, forming the DNA backbone
seperate DNA polymerase moelcules work on each othe strands in opposite directions, ensuring both strands built at the same time
initiation of DNA replication
particular nucleotides in the origin of replication region that are very concentrated with adenine and thymine which are bases with only 2H bonds between, so they r easier to break(less energy needed)
In eukaryotic cells, there r multiple origins of replication
pre-replication protein complex is an enzyme that binds to these areas to help separate the two strands
forms a replication bubble when the nucleotides are separated. The single-stranded binding protein (SSBP) binds to the vulnerable nucleotide strands to protect the parental strands from nucleases(enzymes) and prevent them from reannealing.
helicase unwinds the DNA at both replication forks, requiring a ton of DNA
totpisomerases
fixes the supercoils created as helicase works
what is a polymerase chain reaction, function?
lab process used to amplify and separate certain DNA sequence, creates millions of copies form just a few original DNA molecule, useful for scientist doing analysis and sequencing etc.
process of PCR
occurs in a thermal cycler
need: DNA primer(more stable and easier to synthesise), DNA sample, nucleotides, taq polymerase, mix buffer and PCR tube
1. During denaturation (around 94-96°C), the
double-stranded DNA template is heated to
separate the two strands by breaking hydrogen
bonds between base pairs
2. In the annealing step (50-65°C), short DNA
sequences called primers bind to complementary
sequences on each strand, marking the start and
end points for amplification.
3. Finally, during extension (around 72°C), Taq
polymerase synthesizes new DNA strands by adding
nucleotides to the primers, creating two complete
copies of the target sequence.
—> cycle repeated 25-35x
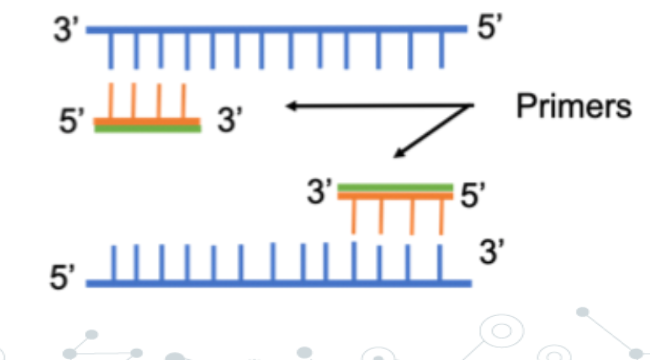
primer
Primers are short, single-stranded DNA sequences* (typically 18-25 nucleotides long) that are complementary to the beginning and end of the target DNA sequence. A primer is needed because all DNA polymerases can only add nucleotides to a pre-existing strand with a free 3'-hydroxyl (-OH) group, it cannot start from scratch.
Cells use RNA primers for DNA replication; PCR uses DNA primers because they are more stable and easier to synthesise.
taq polymerase
Taq polymerase is a heat-stable DNA polymerase originally isolated from the Thermus aquaticus(bacterium which lives in hot springs). This enzyme is crucial because it can withstand the high temperatures used in PCR without denaturing, unlike regular DNA polymerases that would be destroyed by heat.
gel electrophoresis
a technique used widely in the analysis of DNA, RNA, and proteins
During electrophoresis,the molecules are separated with an electric current according to their size or mass and their net (overall) charge.
This separation occurs because of:
The electrical charge molecules carry: Positively charged molecules will move towards the cathode (negative pole), whereas negatively charged molecules will move towards the anode (positive pole), e.g. DNA is negatively charged due to the phosphate groups and thus, when placed in an electric current, the molecules move towards the anode.
The different sizes of the molecules: Different sized molecules move through the gel (agarose for DNA and polyacrylamide for proteins) at differentrates. The tiny pores in the gel resultin smaller molecules moving quickly, whereas larger molecules move slowly
The type of gel: Different gels have different sized pores that affectthe speed at which the molecules can move through the gel
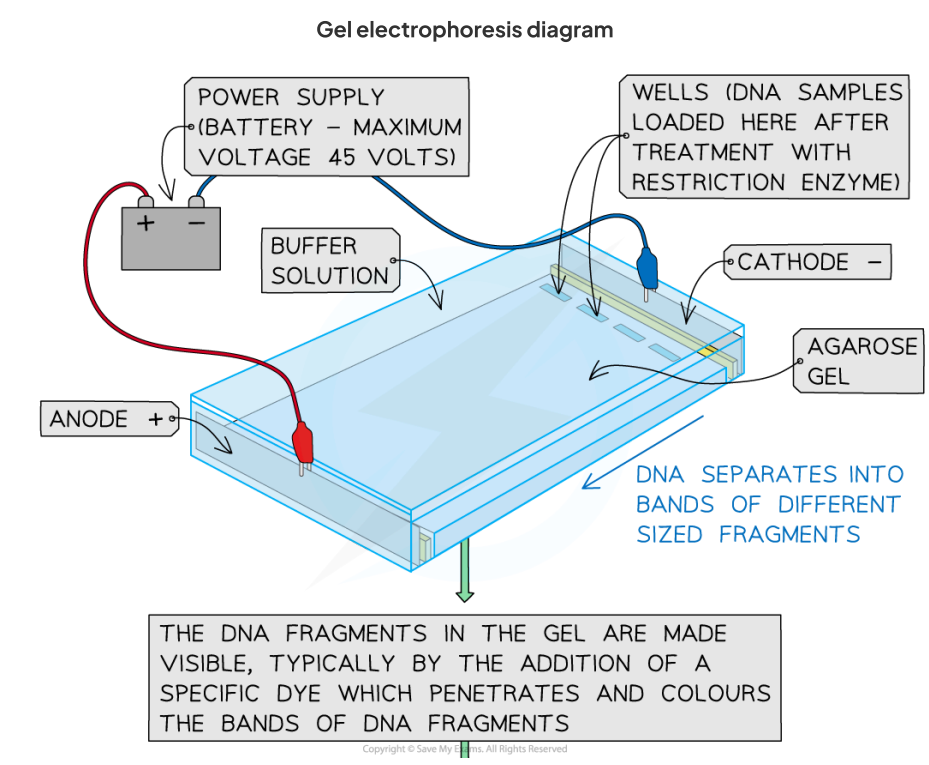
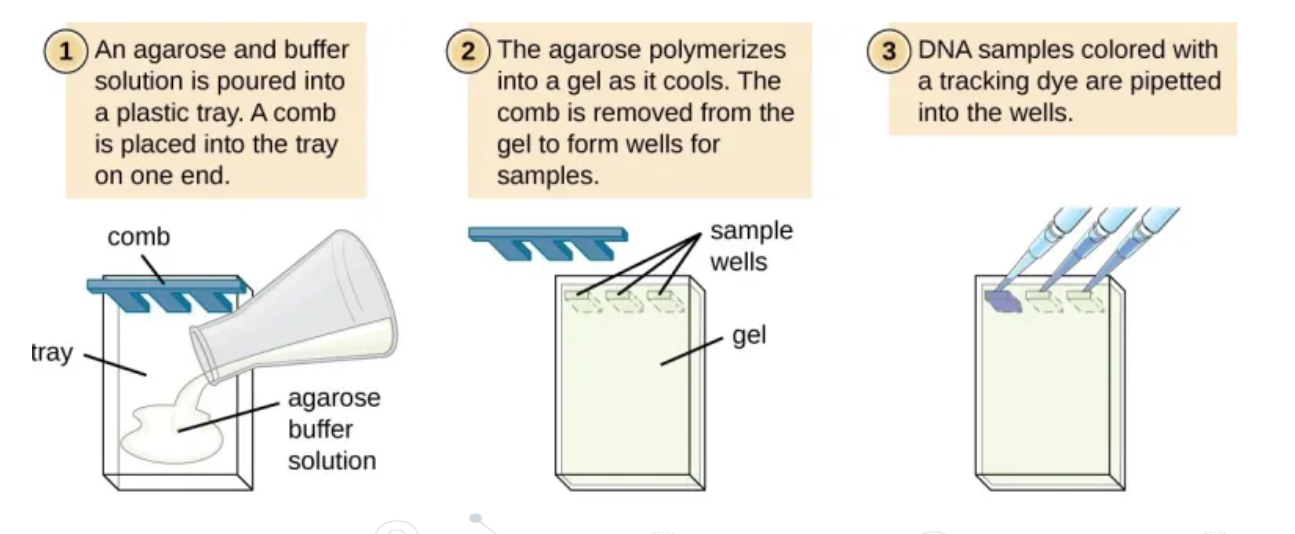
process of gel electrophoresis
Create an agarose gel plate in a tank. Wells (a series of small rectangular holes) are cutinto the gel at one end
Submerge the gel in an electrolyte solution (a salt solution that conducts electricity) in the tank
Load (insert)the DNA fragments into the wells using a micropipette
Apply an electrical current to the tank. The negative electrode must be connected to the end of the plate with the wells as the DNA fragments willthen move towards the anode (positive pole) due to the attraction between the negatively charged phosphates of DNA and the anode
DNA fragments with a smaller mass (i.e. shorter DNA fragments) will move faster and furtherfrom the wells than the largerfragments
The fragments are not visible so must be transferred onto absorbent paper or nitrocellulose which is then heated to separate the two DNA strands
Probes are then added to develop a visual output, either: A radioactive label (eg. a phosphorus isotope), which causes the probes to emitradiation that makes the X-ray film go dark, creating a pattern of dark bands and a fluorescent stain or dye (eg. ethidium bromide), which fluoresces (shines) when exposed to ultraviolet(UV) light, creating a pattern of coloured bands
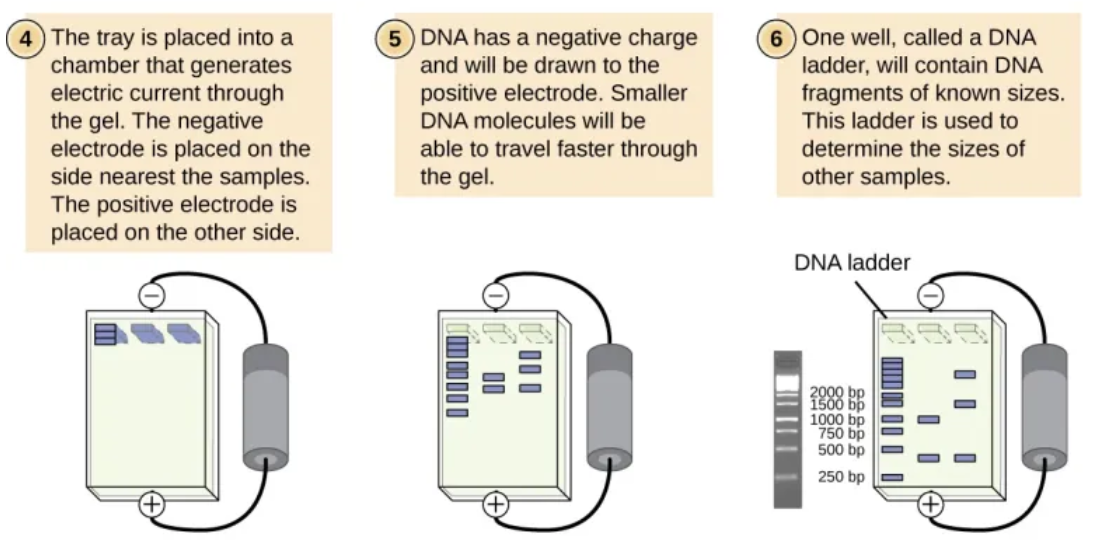
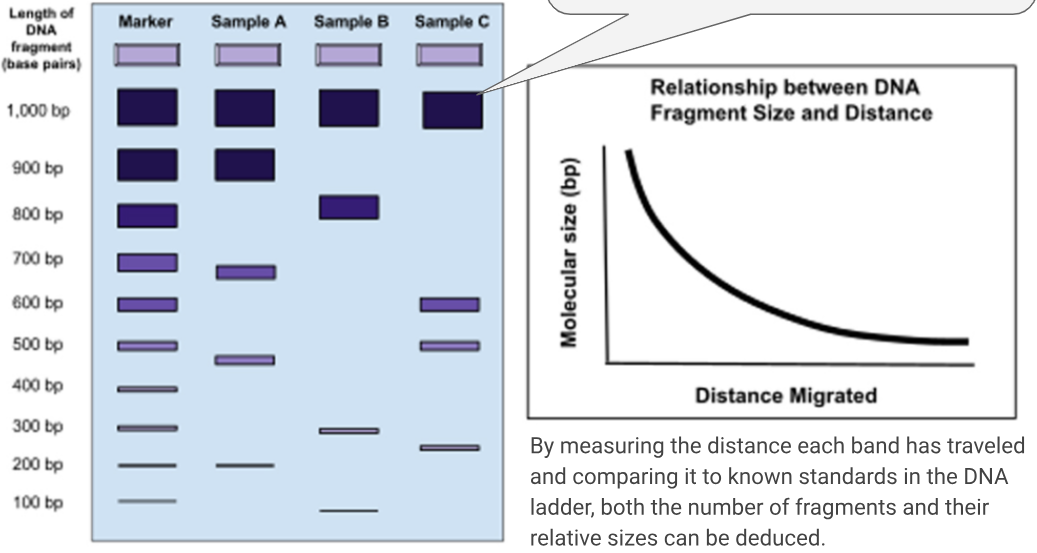
what do the DNA fragments mean on the elctrophoretogram?
The number of bands visible on the gel after staining corresponds to the number of different-sized DNA fragments in each sample. By measuring the distance each band has travelled and comparing it to known standards in the DNA ladder, both the number of fragments and their relative sizes can be deduced.

amplify
Single-stranded binding proteins
keep the separated strands apart whilst the template strand is copied, keeps them from reannealing, and protects the vulnerable strands from nucleases(nasty enzymes)
DNA primase
generates a short RNA primer on the template strands Providing an initiation point for DNA polymerase III to add new nucleotides
how is gel electrophoresis useful?
DNA profiling:
paternity tests
forensic investigations
mass disaster victim identification (identifying remains)
wildlife conservation (tracking endangered species populations)
archaeological studies (analyzing ancient DNA), medical diagnostics (detecting genetic disorders and infections)
food safety testing (detecting contamination), research applications (studying gene expression and mutations)
forensic sciences
DNA profiling
Every person (apart from identical twins) has repeating, short, non-coding regions of DNA (20 to 50 bases long) that are unique to them calledVNTRs (Variable Number Tandem Repeats)
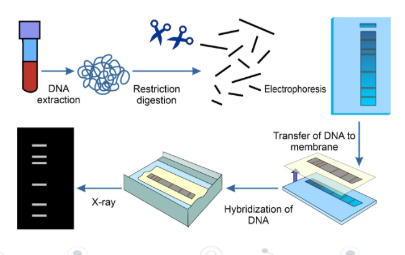
DNA profiling involves using gel electrophoresis to separate VNTR fragments according to length to create a pattern of bands that’s unique to every individual, sometimes called the genetic fingerprint To create a DNA profile from the DNA being tested scientists complete the following in sequence:
1. Obtain the DNA, which can be extracted from the root of a hair, a spot of blood, semen or saliva
2. Increase the quantity of DNA by using PCR to produce large quantities ofthe required fragment of DNA from very small samples (even just one molecule of DNA or RNA).
3. Use restriction endonucleases to cutthe amplified DNA molecules into fragments
4. Separate the fragments using gel electrophoresis
5. Add radioactive or fluorescent probes that are complementary and therefore bind to specific DNA sequences
6. X-ray images are produced or UV lightis used to produce images ofthe fluorescentlabels glowing
7. These images contain patterns of bars (the DNA profile) which are then analysed and compared
paternity tests
DNA profiles ofthe mother and child are compared, along with the profile ofthe alleged father (all three are needed). Patterns of bands are compared on allthree genetic profiles and any band that appears in the child's profile must show in either the mother's or father's profiles; if not,the alleged true father is a different man
restriction enzyme
act as molecular scissors, cutting DNA at specific base sequences
(recognition sites), the resulting DNA fragments are then separated by size using gel electrophoresis, creating a distinct banding pattern that reveals genetic
differences between individuals.
directionality
Similar to transcription and translation, DNA replication must occur in the 5’ to 3’ direction DNA polymerase only works in a 5’ to 3’ direction, adding nucleotides to the 3’ end of a strand of nucleotides DNA nucleotides have a phosphate bonded to the 5’ carbon ofthe deoxyribose sugar
When DNA polymerase adds a new nucleotide to extend the DNA strand, the 5’ phosphate group ofthe incoming DNA nucleotide bonds to the free 3’ -OH group on the growing strand
experiment proving DNA replication is semi conservative
Crick and Watson made a hypothesis about how DNA copies during cell growth, proposed a semi-conservative model, but had not provided the evidence
Meselson and Stahl’s results gave the necessary support for Crick & Watsons' hypothesis of semi-conservative replication of DNA

Meselson and Stahl’s experiment
Meselson and Stahl conducted their famous experiments on DNA replication using E. coli bacteria as a model system.