Ch. 13 + 16: Bacterial Genome and Genome Variations
1/205
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
206 Terms
What is DNA the template for
DNA replication and transcription

replication
DNA → DNA
create 2 identical copies of DNA molecule
DNA polymerase
DNA template read 3’ → 5’
DNA synthesized 5’ → 3’
transcription
DNA → RNA
copying DNA sequence into mRNA molecule
RNA polymerase
DNA template read 3’ → 5’
mRNA synthesized 5’ → 3’
translation
RNA → Protein
use mRNA sequence to build protein
ribosome
mRNA codons read 5’ → 3’
polypeptide synthesized N → C
info on maturation at N terminus
at least one sulfur containing AA in sequence
replication, transcription, and translation in EUKARYOTES
replication and transcription: nucleus
translation: cytoplasm
replication, transcription, and translation in PROKARYOTES
replication, transcription, and translation occur in cytoplasm
What carries out DNA replication
DNA polymerase - few errors; 1 in a million
What carries out transcription
RNA polymerase (enzyme that synthesizes RNA from DNA)
What is mRNA the template for
proteins
What carries out translation
ribosomes
What does reverse transcriptase do
RNA into DNA (specifically cDNA)
identified from viruses
can you turn protein into RNA
no
degeneracy
Griffith
transformation test
demonstrated that non-virulent bacteria can be transformed into virulent forms
What did the transformation test show
the change of nonvirulent organisms into virulent ones via transformation
combining dead versions of bacteria with living versions of nonpathogenic bacteria → pathogenic bacteria
What traits did the transformation show
S strain and R strain
S strain
smooth, pathogenic, has capsule
R strain
rough, not pathogenic, no capsule
What did Avery-Macleod-McCarty do
identify DNA as the transforming factor (when DNA was destroyed, transformation did not occur, not pathogenic)
treating type S DNA extract with DNase didnt lead to transformation of Type R cells
What did Hershey-Chase experiment prove
that DNA is the genetic material because only the phage DNA entered the cell
utilized radioactive elements (35S and 32P) and bacteriophage to show that DNA is genetic material
What did the Hershey-Chase experiment use as a model
bacteriophage T2 (DNA virus)
Details of the Hershey-Chase experiement
DNA was labeled with 32P
Protein coat was labeled with 35S
Only the labeled DNA entered the cell
Why did only DNA enter the bacterial cell
because phages leave the coat outside
What would’ve happened if the Hershey-Chase experiment was conducted with a mammalian virus
both the protein coat and DNA would’ve entered the cell
What are nucleic acids
polymers of nucleotides
What are nucleic acids linked by
phosphodiester (covalent) bonds
How do DNA and RNA differ
RNA: Uracil, OH on C2, single-stranded
DNA: Thymine, H on C2, double-stranded
strand structure of viruses (nonliving)
can be either double stranded RNA or single-stranded DNA
nucleoside vs nucleotide
nucleoside: sugar and nitrogenous base
nucleotide: sugar, nitrogenous base, phosphate
may be mono, di, or triphosphate
What are the purines
adenine and guanine (PURe As Gold)
double ring
Pyrimidines
cytosine, uracil, and thymine
single ring
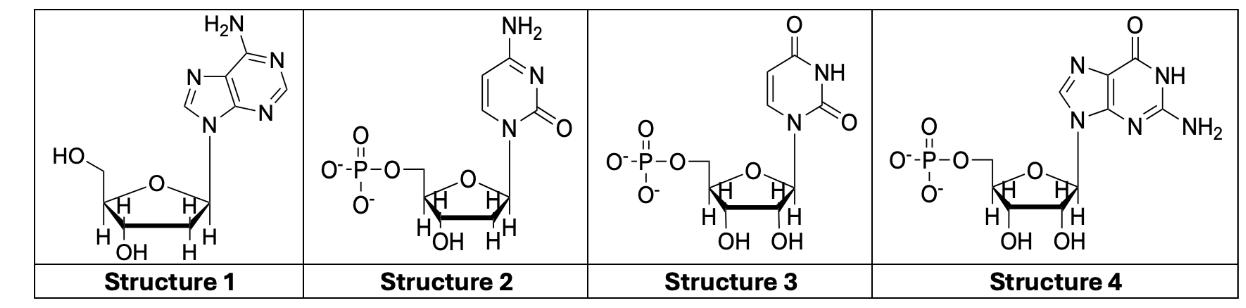
identify nucleoside vs nucleotides / DNA vs RNA / purine vs pyrimidine
1: DNA, nucleoside, purine
note the H on carbon 2 (DNA), and lack of phosphate
double ring: purine
2: DNA, nucleotide, pyrimidine
note the H on carbon 2 (DNA), and presence of phosphate
single ring: pyrimidine
3: RNA, nucleotide, pyrimidine
note the OH on carbon 2 (RNA), and presence of phosphate
single ring: pyrimidine
4: RNA, nucleotide, purine
note the OH on carbon 2 (RNA), and presence of phosphate
double ring: purine
What holds together the double-stranded helix
H-bonds
DNA strands are…
anti-parallel and complementary
what direction is DNA read
3’ to 5’
which is the leading and lagging strand of DNA
leading: 3’ (continuous)
lagging: 5’ (discontinuous)
Okazaki fragments
discontinuous pieces of newly synthesized DNA that form on the lagging strand during DNA replication
What direction do nucleotides get added to DNA
5’ to 3’
how to describe DNA replication
semi-conservative - one old and one new strand
how many times does DNA replicate a genome during a replication cycle
once
where does phosphate bind on DNA
carbon 5
What holds together the back bone of DNA
phosphodiester (covalent) bonds
How many bonds do A-T have
2 H-bonds
what kind of area does RNA polymerase bind in
areas that are A/T rich
weaker bonding makes the area easier to unwind and separate for transcription initiation
broken apart by heat and enzymes
breaking apart with covalent bonds requires a lot of energy
How many bonds do C-G have
3 H-bonds
Major Groove
contains the most information and you can read the nucleotides
What is a ribozyme
an RNA enzyme
carries genetic information
Most RNA molecules are…
single stranded
tertiary structure gives it function
contains both purines and pyrimidines
used as template for DNA
What virus contains double-stranded RNA
rotavirus
What are polypeptides built from
amino acids (monomer)
What bond holds together polypeptides
peptide bonds (covalent)
How are polypeptides synthesized
Amino-terminus (N) to carboxyl-terminus (C)
what gives polypeptides function
3D structure
called a protein once it is properly folded and functional
proteins are made up of how many polypeptides
single or multiple
how are proteins denatured
by pH and temperature; breaks nonvocalent bonds
loses secondary, tertiary, quaternary structure
4 levels of structure of proteins
Primary, Secondary, Tertiary, and Quaternary
Primary proteins
covalent bonds between amino acids
per HW: a sequence of a chain of AA
Secondary proteins
alpha helix and beta sheets, H-bonds
structures formed via hydrogen bonding between carbonyl and amino group of the peptide backbond (alpha helices and beta sheets)
Tertiary proteins
noncovalent bonds + disulfide bridge (covalent)
3D folding driven by side chain interactions
Quaternary proteins
Noncovalent bonds + disulfide bridge
ex. Insulin
structure consisting of multiple subunits coming together
alpha carbon is attached to
a hydrogen, R group, amino group, and carboxylic acid group
categories of amino acids
polar, nonpolar, or charged
important AA to note
methionine (AUG) and cysteine (UGU, UGC)
What does methionine have
sulfur to identify proteins
it is the 1st amino acid added to polypeptide chain (AUG)
every polypeptide should have a methionine
What does cysteine have
sulfhydryl group
What allows disulfide bond/bridges to be made
sulfhydryl group on cysteine
disulfide bridge is between 2 cysteines in the same polypeptide
tertiary structure
disulfide bridge is between 2 cysteines in different polypeptides
quaternary structure
what shape is DNA in most bacteria
circular
what direction is bacterial DNA replication
bidirectional from oriC to ter site
bacterial DNA replication consists of
two replication forks: bidirectional replication from oriC
Works in both directions from OriC → forks meet at Ter
single replicon: one chromosome; one beginning and end for replication
DNA pol III, single oriC site, topoisomerase
what is a replisome
complex made of polymerase, nucleotides, and topoisomerases moving in both directions; carries out DNA replication
DNA polymerase III: adds NT in 5’ → 3’ direction
dNTP (deoxynucleoside triphosphates): raw materials (A, T, G, C building blocks) added to growing DNA strand
xerCD: separates duplicated chromosomes at the end of replication (ter)
topoisomerases
Relieve supercoiling ahead of the replication fork by cutting and rejoining DNA
bacterial RNA polymerase structure
Core enzyme
5 subunits
Sigma factor (σ)
core enzyme + sigma factor = holoenzyme (binds to DNA at promoter)
core enzyme
responsible for elongation phase
Made of 5 subunits: two α, β, β′, ω
β and β′: bind to DNA and form the catalytic center
allows RNA polymerase to bind to DNA
makes RNA but can’t start alone
sigma factor
like a targeting system
recognizes NT sequence
finds the promoter on DNA and start transcription
After initiation, sigma falls off — not needed for elongation or termination
Most common one: σ⁷⁰ (used in normal growth conditions)
holoenzyme
core enzyme + sigma factor = holoenzyme (binds to DNA at promoter)
functional transcription started
what is a promoter
upstream DNA region;
recognized by RNA polymerase with help of sigma factor → strands separate and RNA synthesis begins
not transcribed
RNA polymerase reads DNA 3′ → 5′, synthesizing mRNA 5′ → 3′
promoter features
-10 region: A/T rich (easy to separate d/t 2 H bonds)
-35 region: upstream recognition site for RNA polymerase
+1 site: where transcription begins
three types of RNA
mRNA, tRNA, rRNA
mRNA (messenger RNA)
carries message (coding instructions) for protein synthesis
translated into AA
tRNA (transfer RNA)
carries AA to ribosome during translation (protein synth)
has anticodon that pairs with mRNA
rRNA (ribosomal RNA)
components of ribosomes (rRNA + protein)
has secondary hairpin loop structures
are tRNA and rRNA translated
no, they are functional RNAs
their function depends on the NT sequence and 3D shape
in bacteria, transcription has…
a defined start site (promoter)
ends at termination site
genes
basic unit of genetic information
non-protein coding genes
genes that code for tRNA and rRNA; do NOT get translated
what is transcribed together in bacteria
tRNA and rRNA
how are tRNA and rRNA removed
by ribozymes or ribonucleases
structure of rRNA/tRNA gene transcription unit
Promoter – RNA polymerase binding site
Leader – non-coding region before coding region
Coding region – the actual tRNA or rRNA gene
Trailer and Terminator – sequences downstream of coding region
These segments often contain spacer regions - removed during processing
post-transcriptional modification is done by
ribozymes: RNA molecules with catalytic activity (self-cleaving)
made of nucleotides
cut RNA at specific sequences, no protein needed
ribonucleases (RNases): RNA or nucleotides with proteins
cut RNA or RNA-protein complexes
remove spacers if multiple tRNAs are in pre-processed transcript
direction of RNA polymerase reading and synthesis
Reads DNA 3′ → 5′
Synthesizes RNA 5′ → 3′
➤ Always adds new nucleotides to the 3′ end of the growing RNA
protein-coding genes
structural genes that encode polypeptides
via mRNA → translation into polypeptides
anti-sense (template)
DNA template strand that directs mRNA synthesis
complementary to sense and mRNA
sense (coding)
complementary DNA strand that has the same NT sequence as mRNA (except T instead of U)
RNA is a copy of the sense strand but made off the antisense
leader sequence
5’ untranslated region (UTR) of mRNA before coding region
contains shine-dalgarno sequence
Shine-Dalgarno sequence
helps ribosome bind for translation to begin; not translated
polycistronic mRNA
one mRNA that encodes multiple proteins
prokaryotes
monocistronic mRNA
one mRNA encodes one protein
eukaryotes
operon
makes polycistronic mRNA
encodes lac permease
contolled by a single promoter but includes multitude of genes downstream
Contain genes for proteins with similar or same functions
coupled transcription and translation
in prokaryotes (bacteria and archaea)
this is possible because both happen in the cytoplasm