Ch 13: The Genetic Code and Transcription
1/58
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
59 Terms
Central Dogma
DNA —transcription—> RNA —translation—> protein
RNA polymerase
directs synthesis of RNA using a DNA template
5’ —> 3’ direction
no primer required
enzyme uses NTPs
RNA synthesis
mRNA
intermediate molecule between DNA and proteins, carries info for order of amino acids
n(NTP) + DNA tempalte —RNAP—> (NMP)n n(PPi)
reaction of RNA synthesis
best understood so underlies all we know
many features conserved
target of antibiotics
Why start with prokaryotes?
Initiation
Elongation
Termination
Transcription stages
holoenzyme
whole enzyme
σ
RNAP core enzyme lacks…
only 1 RNAP that produces all types of RNA
mRNA not modified after synthesis
genes arranged in polycistronic operons
transcription and translation coupled
Prokaryotic transcription
interacts with DNA and transcription factors to play regulatory role
α E.coli polymerase subunit
nonessential and regulatory
ω E.coli polymerase subunit
bind DNA and synthesize RNA
β and β’ E.coli polymerase subunit
promoter recognition during initiation
not present during elongation
confer specificity allowing different genes to be expressed
σ E.coli polymerase subunit importance
-35 = TTGACA and -10 = TATAAT
E.colli promoter sequences
consensus sequence
a sequence of DNA nucleotides most frequently found at each position and is necessary for particular function
σ70
Major E.coli σ factor
promoter
where RNAP binds to start transcription, upstream of sequences
open-complex
part of initiation where single-stranded bubble created to go from stable, closed complex to active site
RNAP finds and recognized promoter via σ subunit and -35 and -10 sequences
RNAP binds and partially unwinds DNA creating open complex formation
E.coli initiation steps
Promoter regions of many genes were aligned by start site of transcription and found to have similar sequences at set distances
How were consensus sequences discovered?
40 nt/second
RNAP rate (nt/sec)
800 nt/sec
DNAP III rate (nt/sec)
15 aa/sec moving at 45 nt/sec
Prokaryote ribosome rate (aa/sec and nt/sec)
template strand (nonsense strand)
being copied, anti-parallel to RNA
coding/sense strand (makes “sense” when translated)
looks like mRNA
RNAP moves away from promoter with core enzyme synthesizing RNA in 5’ —> 3’ direction
3’-OH = nucleophile to attack α phosphate on incoming nucleotide
σ releases
E.coli Elongation steps
Rho independent and Rho dependent
E.coli termination types
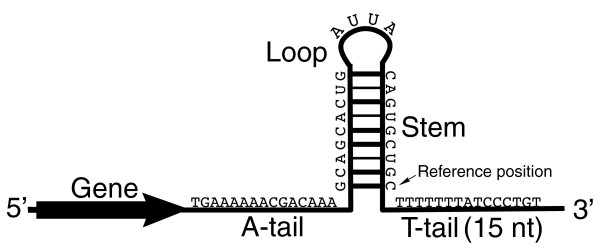
no protein required, based on nucleotide sequence
RNA folds into hairpin with G-C rich stem followed by string of U
rU-rA bonds stronger than rU-dA —> weakest interaction left with DNA causing dissociation
Rho-independent termination
hexamer that recognizes rut base sequence
wraps RNA around itself
uses ATP to pull RNA from DNA
Rho-dependent termination
operon
a cluster of related genes under control of a single promoter
multiple RNAPs
occurs in nucleus
not coupled with translation
transcription occurs on chromatin
requires chromatin remodeling
require processing to produce mature mRNAs
Differences in eukaryotic transcription vs prokaryotic transcription
rRNA
RNA Pol I products
mRNA, snRNA
RNA Pol II products
tRNA, 5S rRNA
RNA Pol III products
12 subunits (10 required)
RNA Pol II subunits
Which cells gene expressed
When gene expressed
How much gene expressed
What do enhancers and repressors regulate?
core promoter elements
required for direct accurate initiation of transcription by RNA Pol II machinery
recognizes promoter and assembles initiation complex
recruits other GTFs and RNA Pol II
General transcription factors (GTPs) roles
GTF (TFIID) recognize promoter sequence and bind
Recruit other GTFs and RNA Pol II
Pre-initiation complex formed
Eukaryote Initiation step
GTFs phosphorylate CTD
machinery disassembles and RNA Pol II leaves
RNA Pol II adds first NTP without 3’-OH
Bridge helix moves DNA and RNA during elongation
mRNA processing
Eukaryote Elongation steps
hnRNA (heterogenous nuclear RNA)
initial mRNA name
unusual 5’-5’ triphosphate linkage
binds to phosphorylated CTD and moves to mRNA
recognized by translation factors and protects against exonucleases
Eukaryotic 5’ 7-methyl-G-cap
introns
regions of the initial RNA transcript that are not found in the mature mRNA
U1 snRNP
spliceosome example
U1 snRNA binds to 5’ splice site of intron via RNA-RNA interaction
U2 and hnRNA branch point (A) bind
U1 and U2 interact
other SnRNPs come in
2’-OH on 3’ branch site attacks phosphodiester bond art 5’ splice junction
3’-OH generated and Lariat loop forms
3’-OH attacks 3’ splice site
Mature mRNA created and Lariat intron degraded
Mechanism of spliceosome splicing
GTP binds to active site in intron
3’-OH on GTP breaks phosphodiester bond at 3’ left exon
New 3’-OH interacts with phosphodiester bond on right exon
intron sliced out and exon joined
Self splicing mechanism
CPF recognizes sequence in mRNA
CPF cuts mRNA and adds polyA tail
Ral1/Rat1 RNase degrades 5’ end moving towards RNA Pol II causing dissociation from DNA
Eukaryote Termination
guide RNAs (gRNAs)
What directs post-transcriptional modifications
changing base or insertion/deletion
What edits can be made post-transcriptionally
TATA box
A short nucleotide sequence 20–30 bp upstream from the initiation site of eukaryotic genes to which RNA polymerase II binds
multiple codons per amino acid
What does it mean that the genetic code is degenerate
each codon is for only one amino acid
What does it mean that the genetic is unambiguous
via degeneracy and chemically similar aa’s sharing similar bases in codon
how does the genetic code buffer against mutations?
AUG
start codon
UAG, UAA, UGA
stop codons
methionine
What does AUG code for?
formylated-methionine (fMet)
Bacteria starting amino acid
wobble hypothesis
hydrogen bonding between the codon and anticodon at the 3rd position is subject to modified base-pairing rules