RRoy 13: Translational (Post-Transcriptional) Control
1/16
Earn XP
Description and Tags
November 21st, 2025
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
17 Terms
is mRNA stability constant across organisms? How do we know?
No, stability varies widely within and between organisms
We can measure stability using half life - the longer the half life, the more stable

What is the benefit of having a short mRNA half-life? (c-myc example)
c-myc is an oncogene that overactivates growth when it is overexpressed
When you have too much of it, it can transform cells
often we need it, but we dont need a lot
Its short half-life of 30 minutes is a regulatory mechanism, as this gets rid of its mRNA shortly after it is produced
that way it gets translated, makes the c-myc but cannot make too many more
What sequence is characteristic to unstable mRNAs?
It was found that regions in the 3’ untranslated region (UTR) of unstable mRNAs had multiple copies of the sequence AUUUA
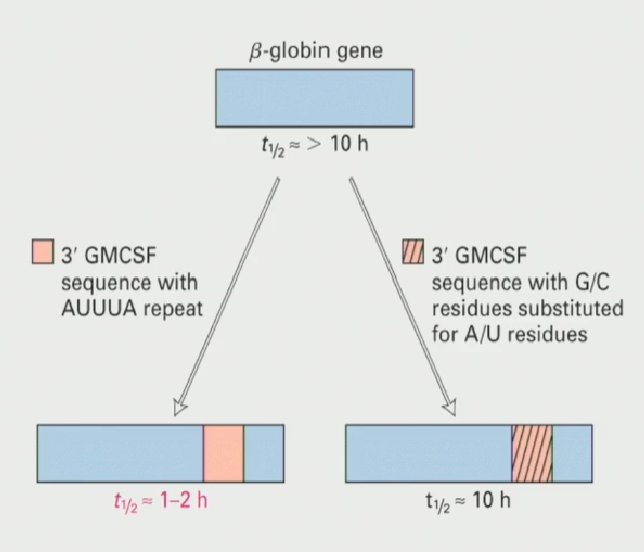
What experiment showed that the AUUUA sequence was sufficient to destabilize mRNA?
Two versions of a cytokine (GMCSF) were inputted into two Beta-Globin genes
One version of GMCSF was rich in AUUUA repeats, and the other had these sequences substituted with G/C residues
It was seen that the GMCSF rich in AUUUA repeats significantly decreased the half life of the beta-globin gene, proving that AUUUA repeats are sufficient to destabilize mRNA
How are the AUUUA sequences sufficient to destabilize mRNA? (How do they begin the process of degradation?)
AU rich sequences are recognized by machinery in the cell that will recruit in degradation proteins that will attack the RNA
Degradation can occur in both directions along the mRNA
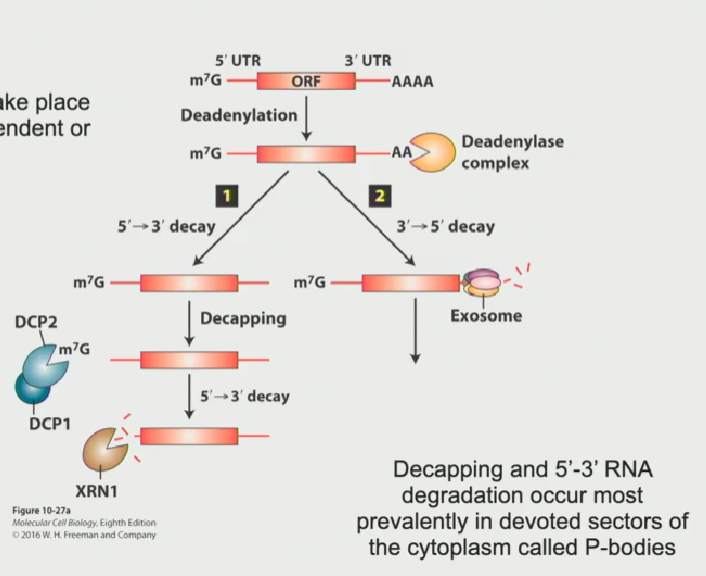
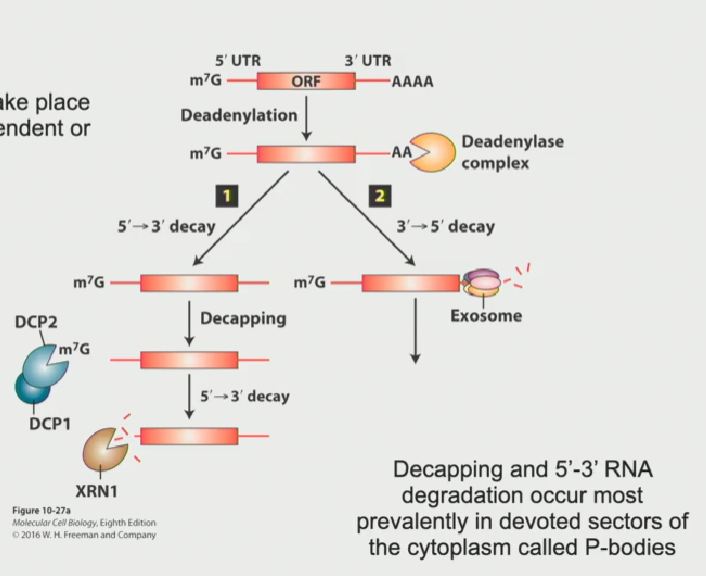
Process of RNA degradation in 3’-5’ direction
Poly-A tail becomes deadenylated by deadenylase (it shortens the Poly-A tail until there isn’t a large enough attachment point for Poly-A binding protein C to attach, leaving the mRNA vulnerable to degradation
Another complex, known as the exosome, attacks the mRNA from the 3’-5’ end
This is the common degradation process in mammalian cells
This method of degradation can take place anywhere in the cytoplasm or in the nucleoplasm
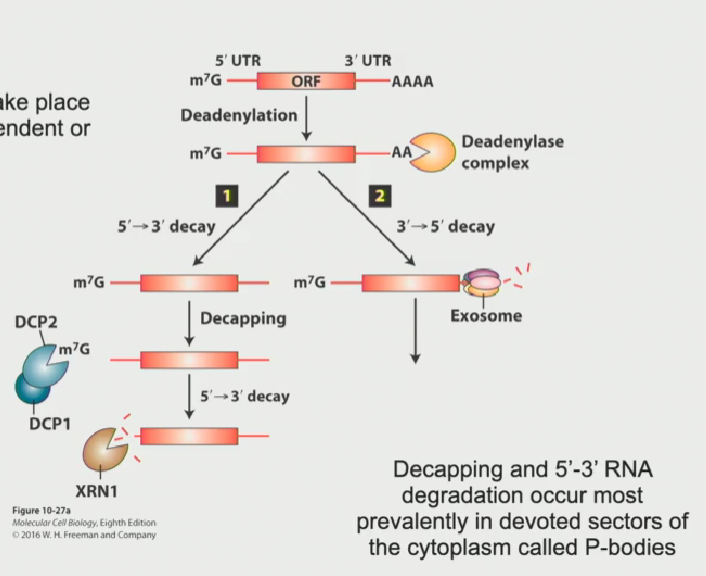
Process of RNA degradation in 5’-3’ direction
7’methylguanalate cap is removed from the 5’ end by the decapping complex (DCP1 + DCP2)
This leaves the 5’ end vulnerable to attack by an exoribonuclease known as XRN1. This will chew the RNA in the 5’-3’ end
This pathway is the most common degradation process in yeast
This method of degradation only takes place in P-bodies (membrane bound organelles meant for degradation)
What is the exosome? What does it include
A structure made to degrade RNAs
RNA will be threaded through the exosome
First, an RNA helicase will remove any secondary structures
ATPase base
Exo-9
Exonuclease activity that chews up the RNA is at the end
Backup endonuclease that sits outside the pore at the end. In case any RNA makes it through, this endonuclease will cut it

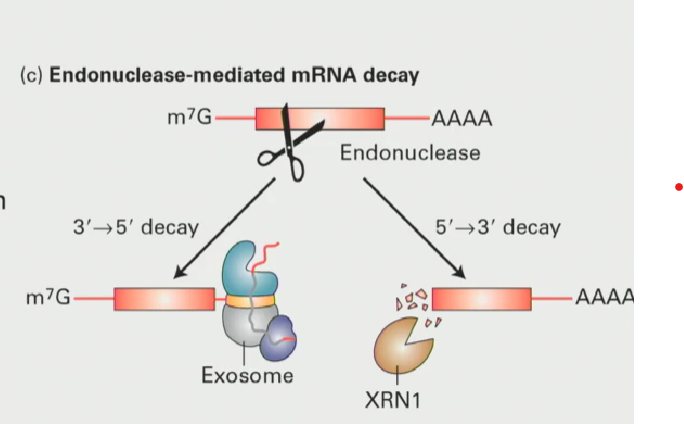
Why is it so deadly for mRNA to be cut by an endonuclease?
This makes a new 5’ end and a new 3’ end: essentially two more spots for degradation to occur in either direction
Regulation of transferrin receptor (iron regulation) purpose + process
Iron is necessary for survival, but toxic if levels get too high
Iron levels must be regulated by the transferrin receptor
Transferrin receptor is critical for bringing iron into the cell so that it can maintain reasonable intracellular levels of iron
If intracellular levels of iron are high, you do not want to bring more iron into the cell so you want to disable the transferrin receptor
Cells evolved a means of doing it by destabilizing transferrin mRNA
Transferrin receptor mRNA has RNA stem loop secondary structures referred to as iron-responsive elements
these elements have AU ruch elements present on their stems
These AU rich elements recruit RNA degradation machinery and, under high iron conditions, the transferrin receptor mRNA is rapidly degraded
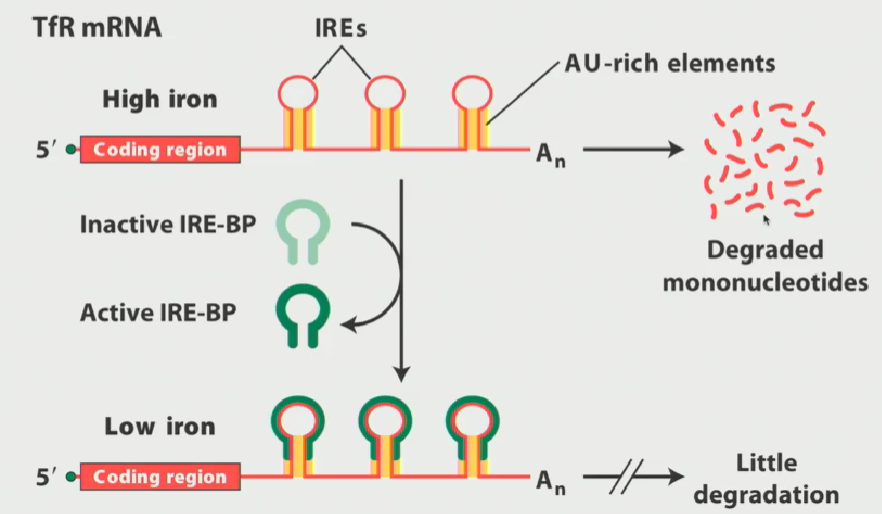
Why are iron-responsive elements important? What are they?
Stem-loop secondary structures present on TfR mRNA (transferrin receptor mRNA) that are rich in AU regions, so they recruit degradation machinery
There is a known RNA binding protein that interacts very specifically with the iron responsive elements (IREs) known as IRE-BP (iron responsive element binding protein)
IRE-BP for regulating iron
a protein that specifically binds to step loop iron responsive elements on TfR mRNA
IRE-BP is in an inactive conformation during high iron situations
When intracellular levels of iron get low, it changes to an active conformation
When it is active, it interacts with IREs covering the stem-loop structure very tightly
This blocks the AU sequences present on the stems from recruiting degradation machinery

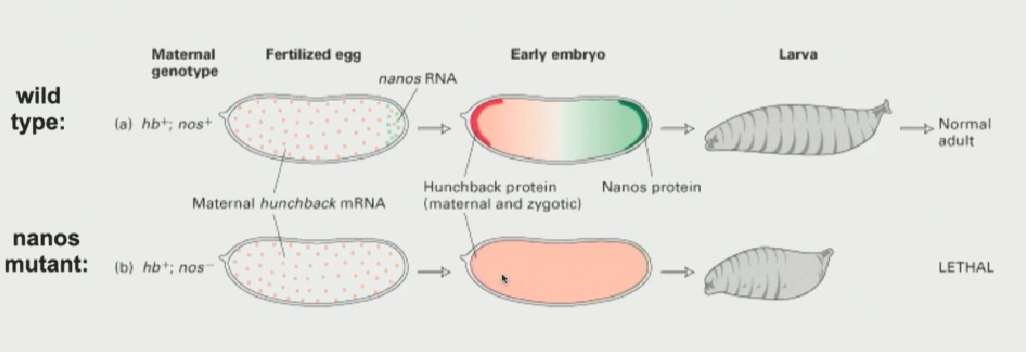
nanos protein / hunchback mRNA (in drisophila)
nanos protein has to sit on the 3’ UTR of the hunchback mRNA. When the protein is not there, the hunchback mRNA can be translated everywhere
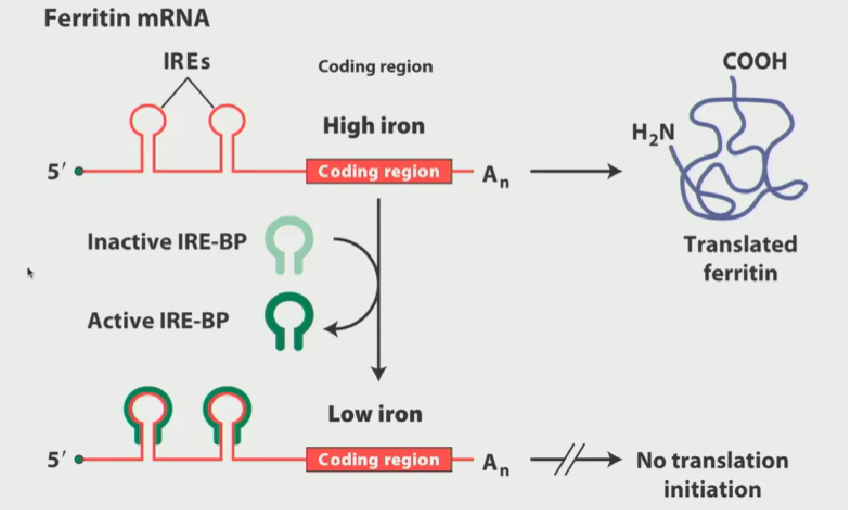
What is ferritin? What does it do?
A protein expressed within cells
Its role is to sequester (bind to) iron so that it is not harmful to the cell
When iron is bound to ferritin, it is no longer dangerous
In high iron conditions, you want a LOT of ferritin
Ferritin regulation explained
Ferritin has 2 IREs (iron responsive elements) in the 5’ UTR (untranslated region) of the mRNA
When the intracellular iron conditions are low, IRE-BP becomes activated and covers the 2 IREs
This strong interaction between the IREs and IRE-BPs blocks the translation of the ferritin mRNA (since we do not need much ferritiin)
C. elegans as a model to study how our body knows when to grow
C. elegans has 4 larval stages, and then adulthood
Every larval stage has unique characteristics
lin-4 is a mutant that made the C. elegans keep repeating larval stage one → never moving onto stage 2 → never becoming an adult
How do animals make the switch between growth stages?
lin-14 is a different mutant that allowed growth to proceed, but there was one version that acted similarly to lin-4
This abnormal lin-14 and lin-4 have to be connected in some way?
Investigators discovered that what was correcting the lin-4 to act normally was an RNA
This mRNA was initially folded into a 62 long sequence with bumps, and was later cut down to a 22 long mRNA called lin-4
One lin-14 unit that didnt act correctly made an mRNA that did not have a UTR
In a wild type lin-14, there are many sites in the 3’ UTR that are imperfect antisense sites to lin-4
lin-4 probably interacts in an imperfect way to these sites in the lin-14 to somehow destabilize it OR block its translation. By doing this, the animal uses this as a trigger to jump into the next larval stage
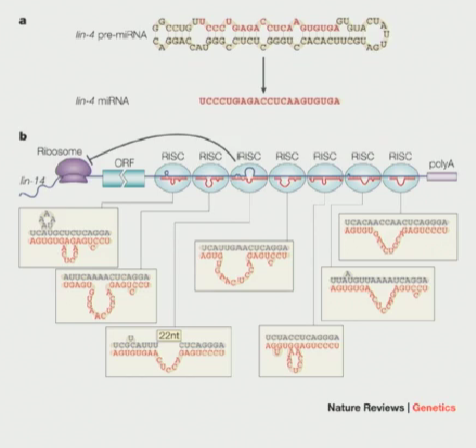
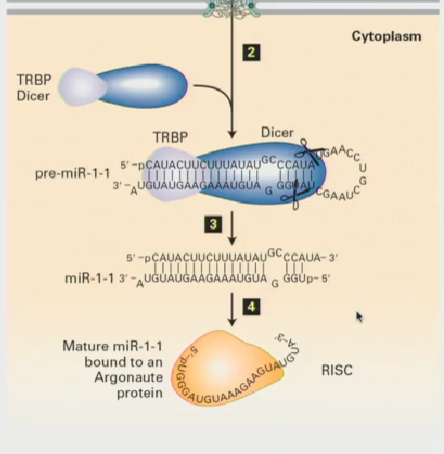
microRNAs (miRNA) & their pathway (Dicer included)
Transcribed by RNA polymerase II & capped
They first form as a primary-miRNA in the nucleus, then are exported via Exportin5 through the nuclear pore complex to the cytoplasm
Once in the cytoplasm, they encounter an RNAse III like enzyme, known as Dicer
Dicer chops the pre-miRNA into small miRNA fragments,which are then incorporated into the miRNA-induced silencing complex (miRISC). These miRNAs guide RISC to target mRNAs, leading to their degradation or inhibition of translation.