Chromosomes
1/42
Earn XP
Description and Tags
L10
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
43 Terms
Statistics that prove we have a lot of long DNA in our cells and body in general
Our genome is about 3 billion base pairs in length
Distance between nucleotide residues 0.34nm
Each cell has about 1 meter of DNA
We have about 37 trillion cells
=> Put together, we have about 37 billion kilometers of DNA in our bodies — Enough to travel from the Earth to Pluto 5 times!!!
DNA is 2nm indiameter
~25 thousandtimes thinner than a hair
What are the two levels of organization of our chromosomes?
chromatin
chromosome
what are the two phases of eukaryotic DNA
interphase (contains G1, S, G2)
metaphase (contains M)
Metaphase
occurs shortly before cell division during mitosis
DNA highly compacted for transmission to daughter cells
this is the X shape of chromosomes we often think of
no transcription
because too compacted
Interphase
all the other stages of the cell excluding metaphase
G1, S, G2
where transcription and DNA replication occur
DNA not as compacted as in M-phase
chromatin
nucleoprotein complex DNA exists as during interphase
is DNA + proteins
equal mass ratio of protein and DNA
can be isolated with an isotonic buffer ( preserves natural structure because has the same salt concentration as the cell’s interior) to be studied
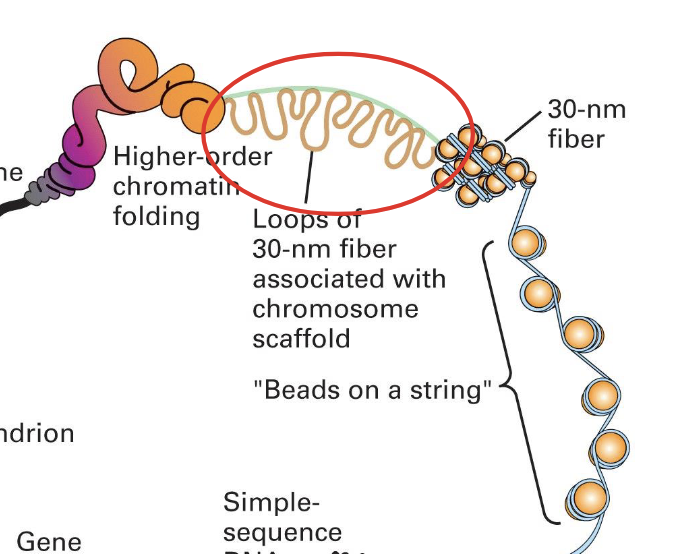
when we remove the salt we see the “beads on a string” structure of the nucleosomes
this is when the chromatin is extended
=> even in its loose state the genetic material is still a highly organized DNA protein
when it is condensed is is about 30nm and fiber)like
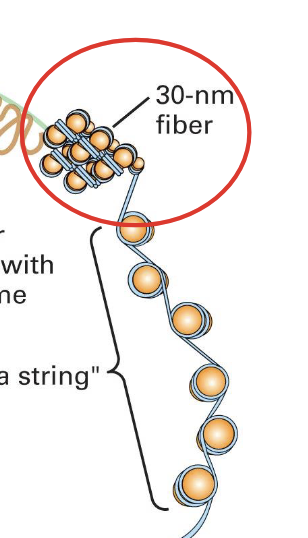
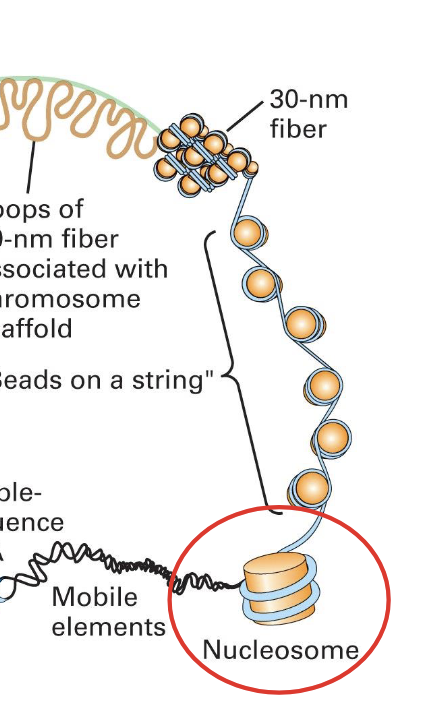
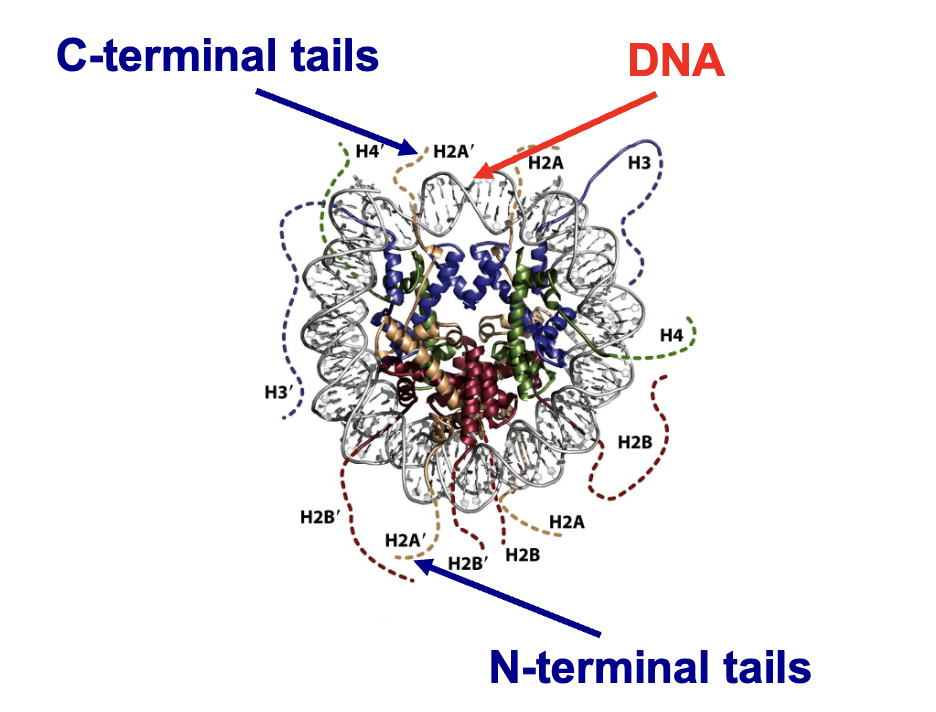
nucleosomes
composed of
histones (proteins)
this is the protein core
DNA
winds around the surface
almost two full turns
separated by linker DNA (10-90 bp in length)
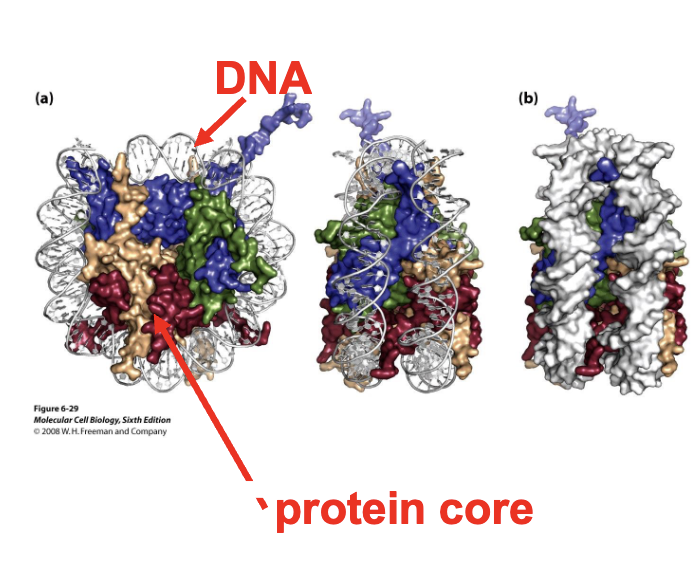
histones
positively charged proteins that interact with the negatively charged DNA
have a core part that wraps itslef with DNA
and tails that stick out of nucleosome and are accesible to other proteins
what does the protein core of a nucleosome consist of
an octamer containing two copies of 4 types of histones
H2A
H2B
H3
H4
what are the two models for the structure of the condensed 30nm chromatin fiber?
Solenoid Model
Consecutive nucleosomes are stacked along a single helical path connected by linker DNA
Two-start Helix Model
aka the zigzag model, nucleosomes are arranged in alternating rows with straight linker DNA connecting non-consecutive nucleosomes in a zigzag pattern
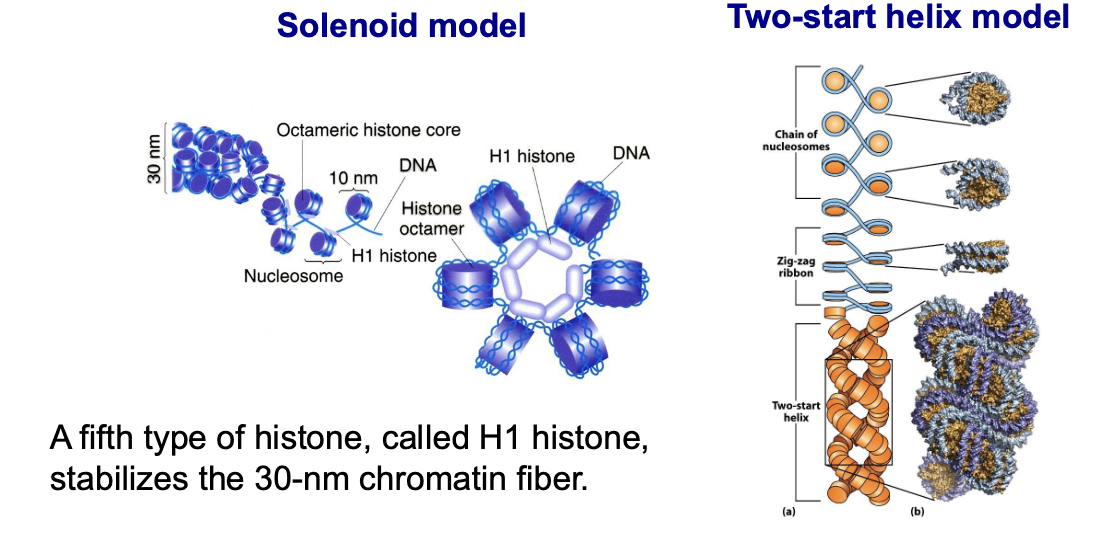
what does the fifth histone H1 do?
it stabilizes the 30nm chromatin fiber
what regulates chromatin condensation?
the modification of histone tails
this is a form of post translational modificaiton
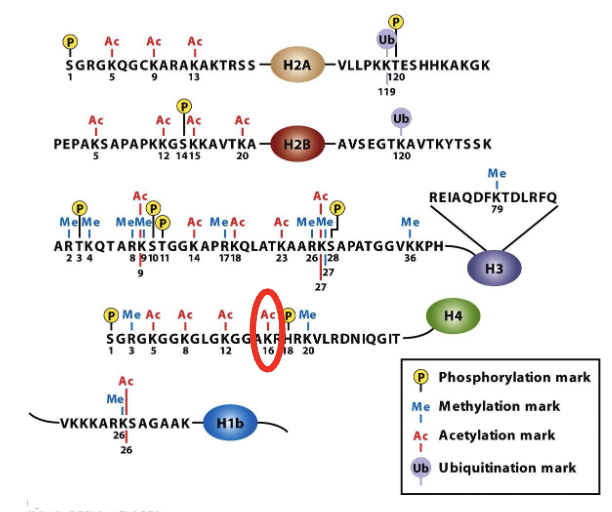
how are histone tails modified?
chemical modificaiton of histone proteins regulate the compaction of chromarin which in turn controls gene expression …by making the DNA more or less accessibel to the transcription machinery
Acetylation of lysine
neutralizes the positive charge of the AA group
other modifications can change the charge of the side chains
e.g.: methylation, phosphorylation and ubiquination (not a mark for degradation in this scenario since just one Ub)
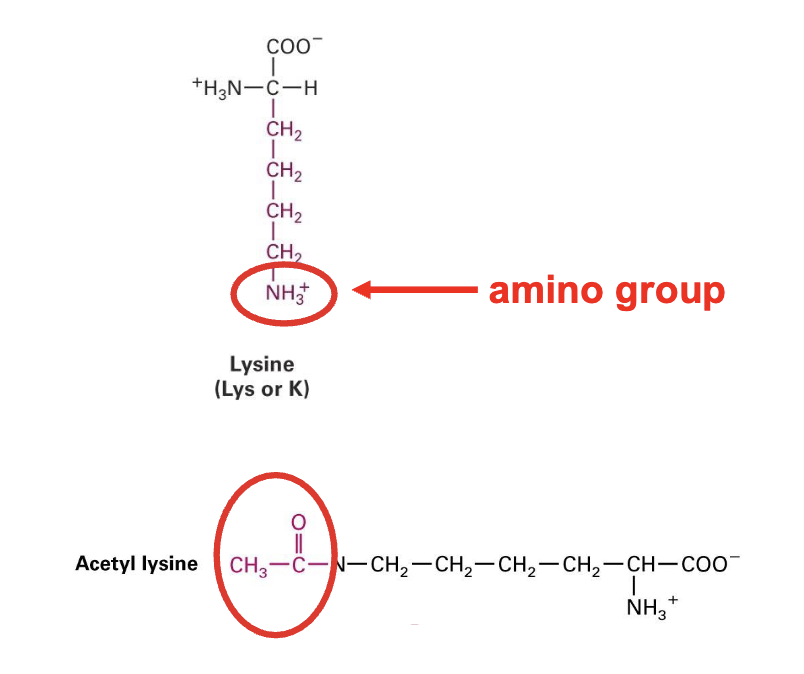
example of a chemical modification: Lysine to Acetyl Lysine
Lysine is a positively charged residue
acetylation removes that charge
leaves us with Acetyl Lysine
In what way is transcription correlated to chromatin condensation
decondensed chromatin
more active
condensed chromatin
not active or less active
What is special about Polytene Chromosomes and what have we studied with them?
There are many parallel identical chromosomes
we often study their giant interphase chromosomes
like that of the Drosophila‘s salivary glands
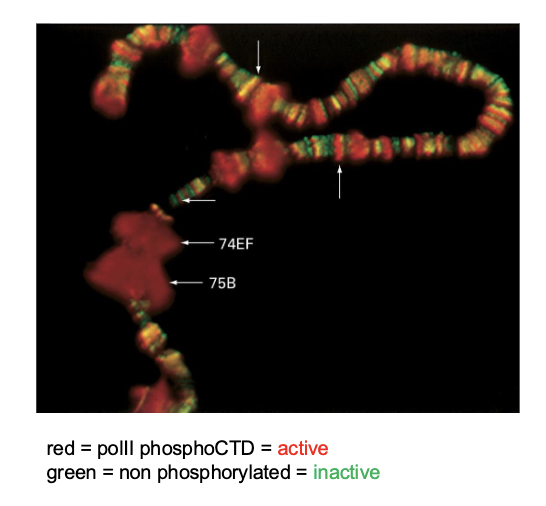
we amplify it to then observe condensed chromatin that presents as dark bands (aka topological domains)
How has polytene chromosome studying shown us that interphase chromatin organization is dynamic?
puffs
show chromatin decondensation
with transcriptional activation — are associated with active form of RNA polymerase II (active transcription)
Heterochromatin
compacted regions
tend to be rich in repetitive DNA
poor in genes
Euchromatin
decondensed regions
gene rich
poor in repetitive DNA
Loops
organized by non-histone proteins (which play other structural roles)
tend to be gene-rich because their primary function is to organize the genome in a way to facilitate gene expression
1-4Mb in length
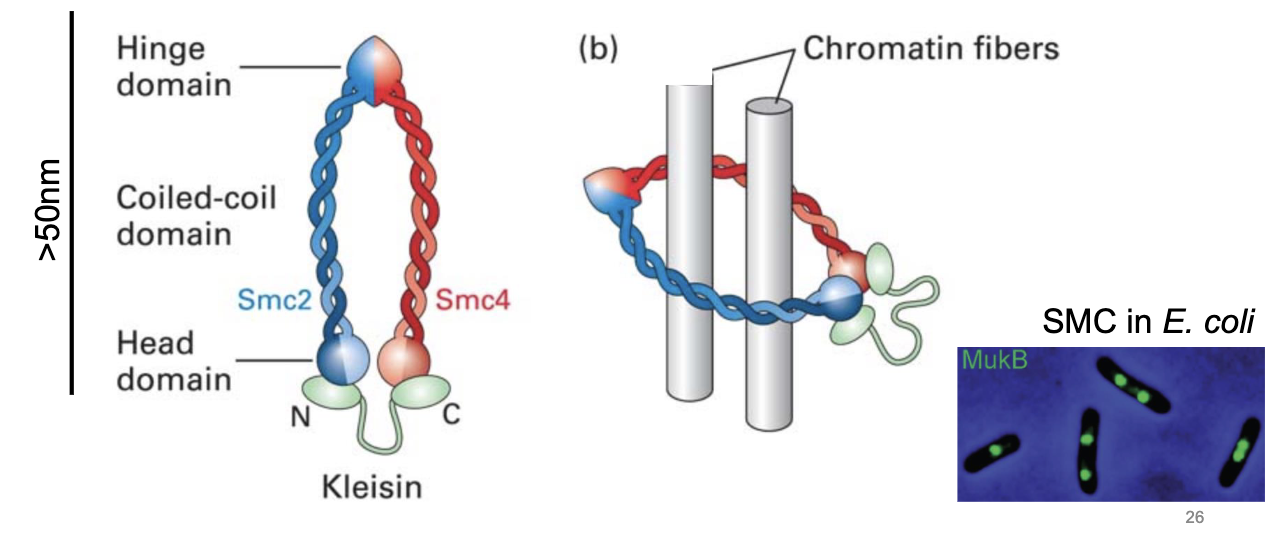
Structural Maintenance Chromosomes (SMC)
an evolutionary conserved family of proteins
we find them in bacteria
they mediate DNA looping
each monomer contains a coiled coil
has a dimer of SMC2 and SMC4
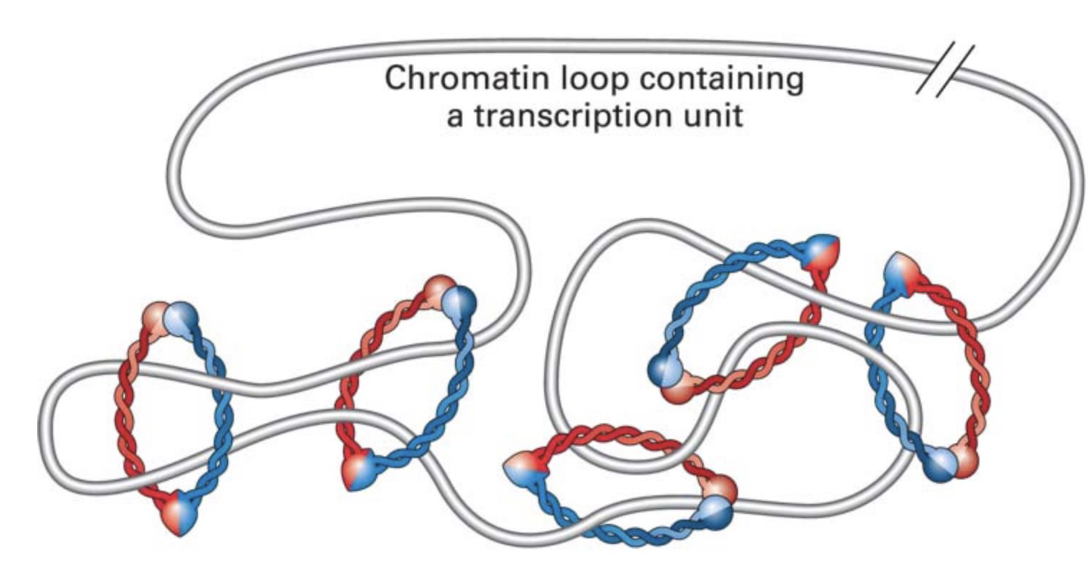
what is the model of loop formation
SMC proteins help hug/encircle DNA to form loops
they slide along DNA/chromatin to make the loops larger or shorter
when do chromosomes occupy most of the nucleus?
during interphase
each one has a particular territory
why does DNA need to be compacted?
for segregation
DNA in our genome has a contour length of 2m so we need to condense them to overcome entangling
how are mitiotic chromosomes organized
as a series of loops around a central core
condensin II forms a central scaffold with loops arounf it
condensin I further compacts these loops into clusters of small nested loops
=> achieves a 10,000 fold compaction of chromatin into linearly organized chromosomes
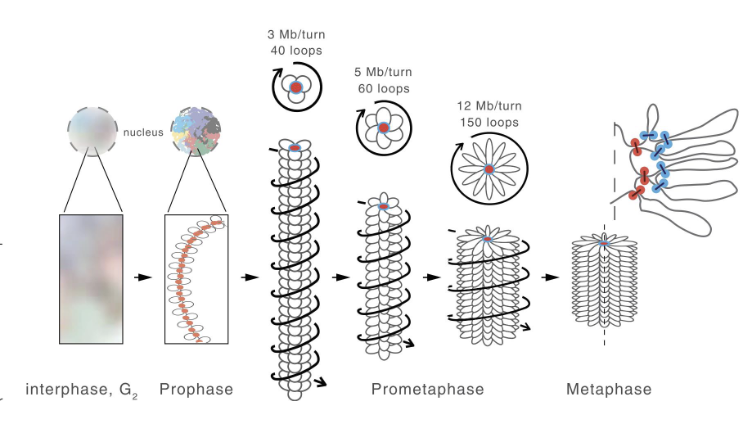
condensins
set of specialized SMCs that condense the chromosomes during metaphase
Condensin I: forms a central scaffold with loops around it
Condensin II: further compacts loops into clusters of smaller nested loops
what are the 3 functional elements required for replication and stable inheritance of chromosomes?
origin of replication
centromere
2 telomeres (ends)
how many origins or replication are there?
multiple
up to 100,000 in the genome
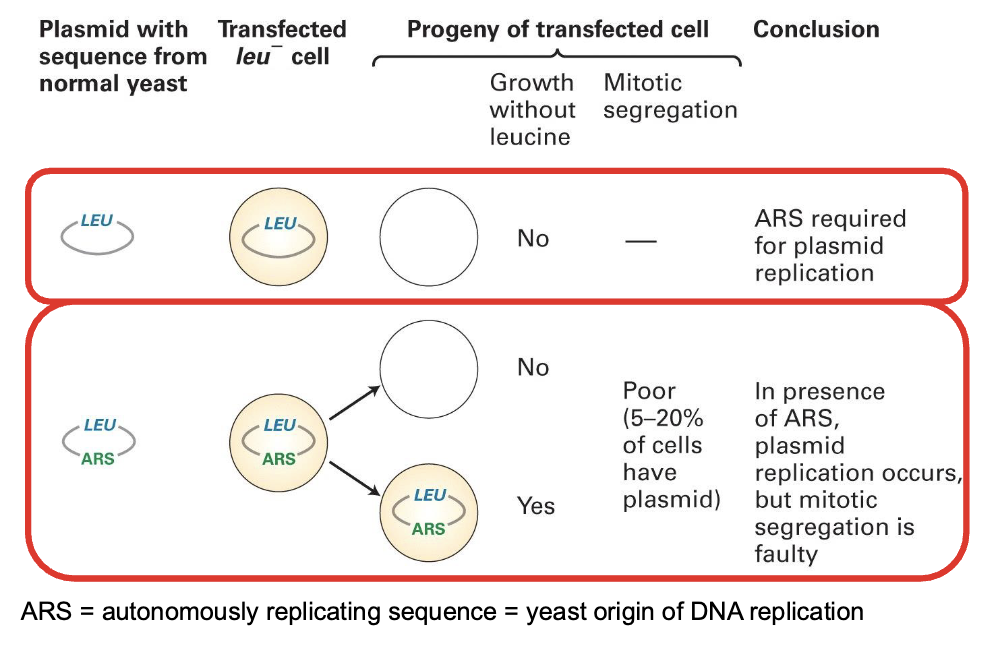
how have experiments done in budding yeasts helped us understand chromosme elements? — yeast origin of replication
In these experiments, we use a circular DNA molecule that carries a gene coding for the enzyme that synthesizes Leucine. Then the cells are grown in environment where there is no leucine so they have t produce their own —> those that can’t die and so we can tell where the gene has been transcribed
ARS (Autonomously Replicating Sequence) is required for plasmid replication
it is the yeast origin of replication
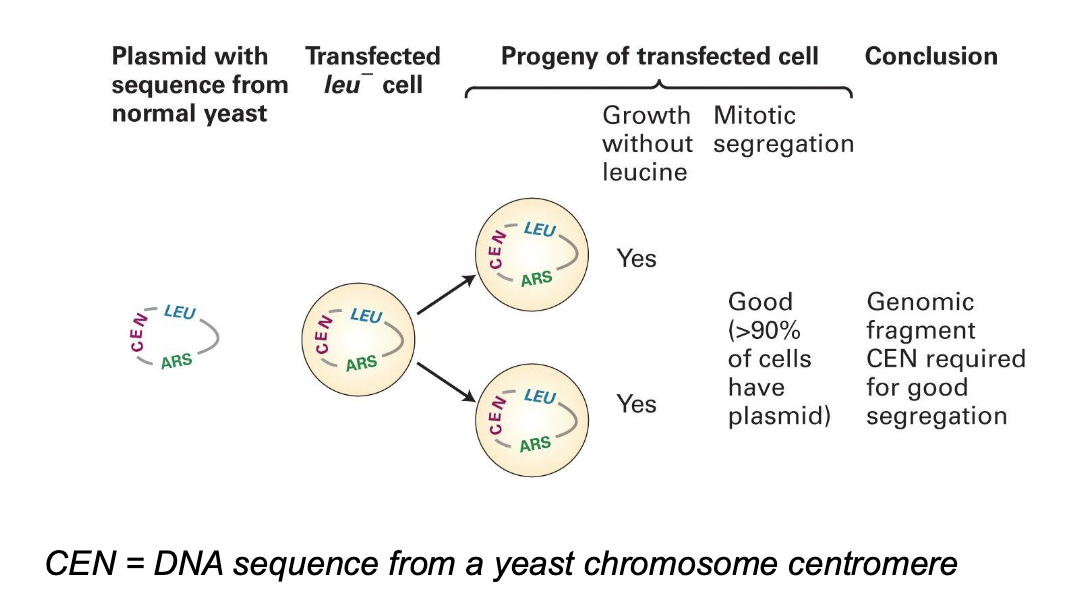
how have experiments done in budding yeasts helped us understand chromosme elements? — yeast centromere
The genomic fragment CEN is the DNA sequence from a yeast chromosome centromere
CEN is required for good segregation
centromere is where genes are linked to microtubules
spindle microtubules
the cytoskeletal structures that pull DNA apart
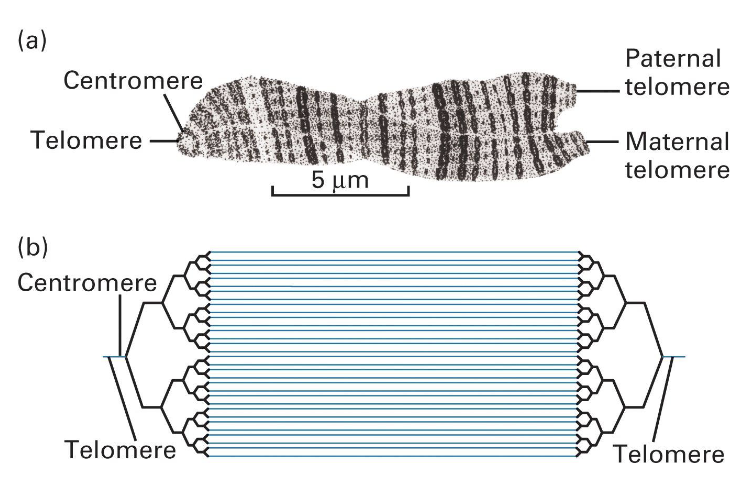
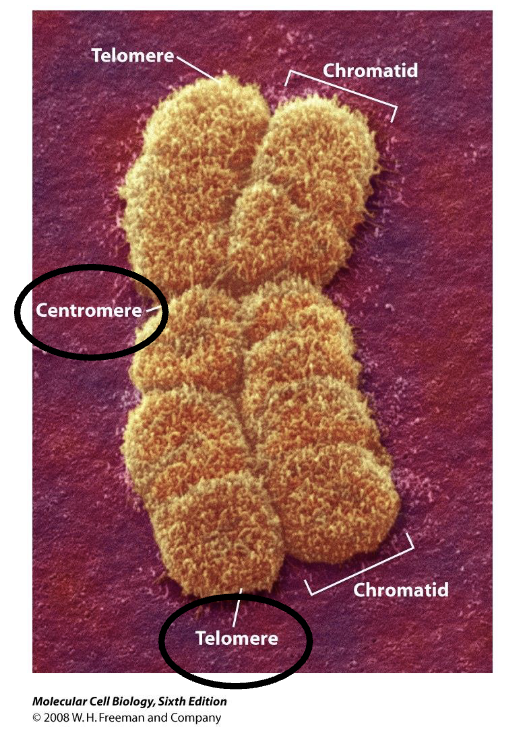
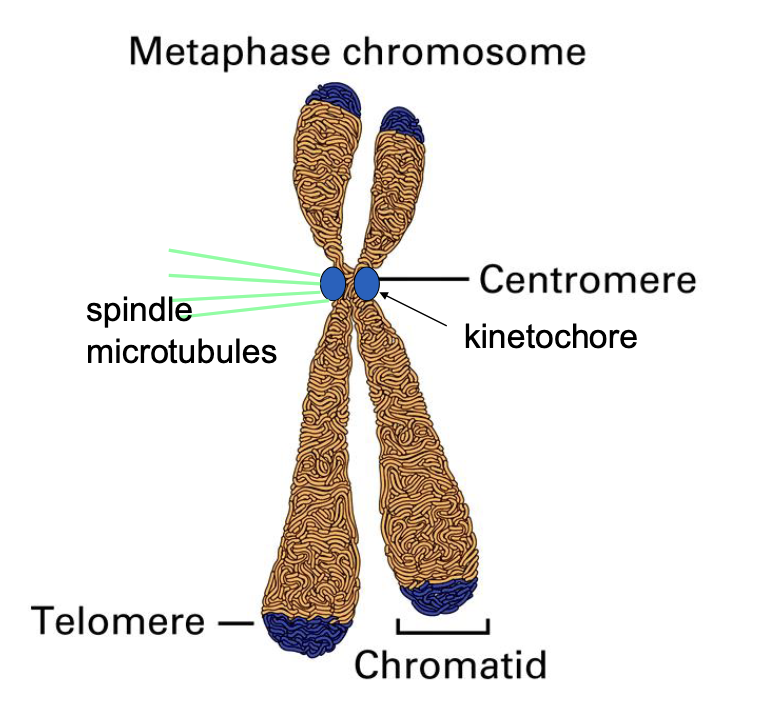
Centromere
constricted region of the chromosome that holds the two sister chromatids together
link to the spindle microtubules
Chromatids
During metaphase, each chromosome consists of two identical DNA molecules called sister chromatids, which were produced during DNA replication.
They are joined together at the centromere and are the most visibly prominent feature of a metaphase chromosome
what are two structures that interact with the metaphase chromsome
kinetochore
a multi-protein complex that assembles on the centromere of each sister chromatid
spindle microtubule
fibers that extend from the spindle poles and attach to the kinetochores. The microtubules are responsible for pulling the sister chromatids apart during anaphase, after the metaphase is complete
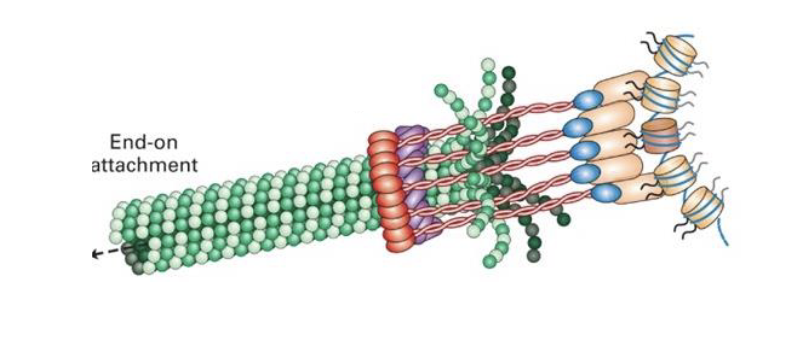
how does the centromere link to the spindle microtubules?
there are sequences common to the various yeast centromeres
contains a nucleosome that includes a centromere-specific histone variant called CENP-A (centromeric protein A)
CENP-A linked to a specialized protein complex called the kinetochore which then connects the centromeres to the microtubules
how have experiments done in budding yeasts helped us understand chromosme elements? — yeast telomere
Here, we use a restriction enzyme to produce a linear plasmid to mimic eukaryotic chromosomes
linear chromosomes lacking TEL sequence for telomere are unstable
linear plasmids containing ARS and CEN behave like normal chromosomes if TEL is added to both ends
Telomere
Protect from exonuclease
Prevent end-to-end fusion
Solve the replication pb faced by linear DNA
by acting as non-coding buffer zones that are progressively shortened with each division, thus protecting the cell's essential genetic information
What is the Telomere Pb?
lagging strand cannot be completed: chromosomes should shorten at the ends in each replication (removal of primers)
chromosome shortening is unsustainable
because at some point will loose an essential gene
telomerase is the solution because it extends the templat to giev primase more template DNA to prime on (preventing the shortening of the ends)
telomerase
a DNA polymerase
can extend telomeres
hence restores chromosome length to overcome lagging strand end-shortening
it is a reverse transcriptase that carries its own template RNA complementary to the DNA repeat
what are telomeres at the DNA level
simple DNA repeat sequences
reverse transcriptase
a DNA polymerase that uses RNA as a template
where is telomerase active
germ cells
stem cells
(not needed in somatic cells because only divide a few tomes so exoisting telomeric repeats are enough)
what is the link between cancer and telomerase
The enzyme is often reactivated in cancer cells
hence could be a target for cancer therapy