MBS 344: Exam 4
1/108
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
109 Terms
What is a non-coding RNA (ncRNA)? What are some examples of ncRNAs in eukaryotic cells?
→ transcribed from the genomic DNA (like mRNA) but is not translated into protein
interact with DNA/RNA to regulate gene expression in euk cells
detect and degrade viral DNA in prok cells
examples? tRNA, snoRNA, rRNA, snRNA
~80% of expressed genes
Compare and contrast long non-coding and small non-coding RNAs. How are they similar? How are they different?
lncRNA: longer than 200 nucleotides and are often used to localize proteins to specific locations on DNA→ bind DNA/RNA using protein & bp interaction
microRNA: shorter than 200 nucleotides and typically bind to mRNA to influence gene expression→ bind DNA/RNA using complementary base pairing
Eukaryotic cells have housekeeping ncRNAs and regulatory ncRNAs. Describe the difference between these two types of ncRNAs and provide some examples of both.
Housekeeping ncRNA: essential for basic cellular functions→ expressed constitutively
snoRNA
tRNA
snRNA
rRNA
Regulatory ncRNA: regulate gene expression by influencing transcription or post-transcriptional events→ expression fluctuated
lncRNA
microRNA
Describe how ncRNAs function as a scaffold and/or guide. Can they bind to DNA, RNA, or both?
lncRNA can form multiple stem loops that make a scaffold→ structure that binds to multiple proteins
lncRNA can act as a guide→ use secondary structures and base pairing to recruit a protein to a specific location in the genome
What is HOTAIR? How does HOTAIR regulate gene expression? How many genes does HOTAIR regulate?
lncRNA that recruits histone-modifying enzymes to their target genes to inhibit gene expression
→ acts as scaffold by binding two different protein complexes using secondary structures
→ guides both complexes to a target gene by binding to DNA at GA-rich regions near target gene
→ once HOTAIR binds DNA, protein complexes alter histone mods at target gene to inhibit transcription
~2200 bases
transcribed by RNA pol II
5’ cap, poly A tail, and undergoes RNA splicing
regulate ~10 genes at chromosome 2
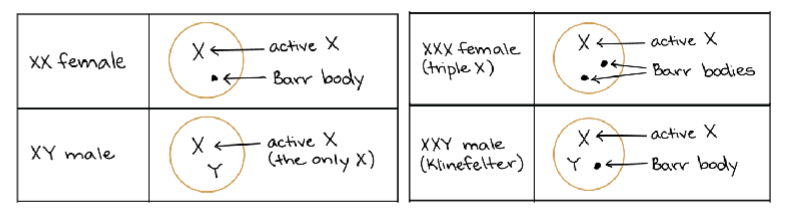
What is X-inactivation? What is a Barr body? What is Xist?
x-inactivation: one of two x chromosome in female cells is inactivated and becomes condensed→ so that both sexes express equal level of genes on x chromosomes (dosage compensation)
Barr body: inactive x chromosome→ heterochromatic so very condensed structure
Xist: lncRNA that only expressed from the inactive X chromosome (Xi) and covers the entire chromosome
Describe some differences between the active (Xa) and inactive (Xi) X chromosomes. Indicating that one is active, and one is inactive is not an appropriate response. Describe why one is active and the other is inactive.
inactive Xi: expresses Xist
histone modifications (H3K27me and H3K9me) promote heterochromatin
active Xa: does not express Xist
histone modification (acetylation and H3K4me) promote euchromatin
How does Xist regulate gene expression? How many genes does Xist regulate?
Xist recruits histone-modifying enzymes that add histone mods to X chromosome and promote heterochromatin
→ regulates hundreds of genes
What are some similarities between Xist and HOTAIR? What are some differences?
same?
same mechanism to regulate gene expression: bind histone-mod enzymes with secondary structure and recruit using base pairing to localize
difference?
genes and number of genes regulated
HOTAIR regulates ~10 genes on chromosome 2
Xist regulates ~hundreds genes on chromsome X
Why is X-inactivation an epigenetic mechanism? Provide at least two reasons.
critical epigenetic process in mammalian development
controls gene expression without altering DNA sequence (histone mods)
heritable— maintained during cell division
Describe why individuals with Turner, Klinefelter, and Triple X syndromes have altered expression from the X chromosome.
deviation from XX or XY causes several health issues
Turner (XO): missing X chromosome (reduced expression of X-linked genes—no escapies
Klinefelter (XXY): x-inactivation shuts doesn’t extra chromosome—incorrect expression bc escapies
Triple X (XXX): x-inactivation shuts down two x chromosome—incorrect expression bc too many escapies
What is DNA fluorescence in situ hybridization (DNA FISH)? What is the purpose of performing DNA FISH?
DNA FISH: lab technique to visualize specific DNA sequence or entire chromosome within a cell
purpose? allows scientist to detect and diagnose genetic diseases
Describe the FISH technique in four steps or less. What is the underlying logic of these steps? Why are cells arrested in metaphase? What are fixed cells?
collect cells arrested in metaphase/interphase and fix cells to glass slide
Metaphase chromosomes are heterochromatic aka condenced, so they are transcriptionally silent
denature DNA in fixed cells to make ss chromosomal DNA
add probe and allow hybridization to specific sequence on chromosome
analyze chromosome in fixed cells with fluorescence microscope— look for probe presence to indicate mutation
What is DAPI? Why is it useful for DNA FISH?
→ flourescent stain that binds dsDNA to allow visualization of ALL chromosomes in cell
What is chromosome painting? Is it considered DNA FISH or RNA FISH? What type of information can chromosome painting provide research scientists or clinicians?
chromosome painting: use multiple DNA FISH probes that bind different locations on the same chromosome
determine if cells have chromosomal abnormalities
ex. analyze X-inactivation
used during interphase (chromosomes not compact)
How does the DNA FISH image change when it is performed with interphase cells (compared to metaphase cells)?
interphase: loosely packed, probes appear as spots on specific DNA regions
regular gene expression→ RNA present
metaphase: condensed, probes appear as chromosome shapes (bands)
irregular gene expression
What is RNA FISH? How is it similar to DNA FISH? How is it different?
RNA FISH: lab technique allows visualization of specific mRNAs within a cells
same?
use probe
use fixed cells
use flour microscope
use DNA probe
different?
RNA FISH does not require denaturation
DNA FISH binds both strands
RNA FISH binds one strand→ requires multiple probes for one mRNA to increase fluorescence
Is RNA FISH used with cells that are in metaphase or interphase? Explain your reasoning.
Interphase! bc metaphase has irregular gene expression so mRNA presence is abnormal while it is normal in interphase.
Describe how DNA FISH and RNA FISH can be used to study X-inactivation. You should be able to predict what DNA FISH and RNA FISH images would look like for patients that have Turner’s, Klinefelter’s, and Triple X syndromes.
DNA FISH: uses gene-specific prove for Xist gene (locate X chromosomes)
RNA FISH: uses probes for Xist mRNA (locate which X chromosomes is silenced)
DAPI: stains all DNA (locate nucleus)
Describe the difference between transcription and translational control (aka post-transcriptional control). How do cells regulate the production of proteins as a form a gene regulation?
transcriptional control
activators bind enhancer OR repressor bind silencer→ influence RNA pol activity
DNA methylation→ inhibit initiation of transcription
lncRNA→ change chromatin structure
translational control
microRNA→ miRNA or siRNA→ interfere with RNA (degrade or silence)
What is RNA interference (RNAi)? Describe how RNAi regulates gene expression.
RNAi: dsRNA processed by cell to induce sequence-specific gene silencing
→ translational control
miRNA: natural
siRNA: experimentally
What is microRNA? How is microRNA different from lncRNA? How is it similar?
microRNA: small non-coding RNAs, too small for secondary structures (do not interact with proteins) rather use base pairing
same? use base pairing
different? no scaffold activity, translational control, much shorter
Describe the biogenesis of miRNA. Where does miRNA come from?
primary miRNA transcribed by RNA pol II in nucleus, processed in nucleus and cytosol, and associated with RISC protein
Describe the miRNA processing using the following terms: double-stranded intermediate, Dicer, primary miRNA, Drosha, and precursor miRNA.
primary miRNA (pri-miRNA) transcribed by RNA pol II and form hairpin
pri-miRNA cut by Drosha in nucleus to form pre-miRNA (ds and ~70 nucl)
pre-miRNA exported to cytosol, Dicer cuts pre-miRNA to make ds-intermediate
ds-intermediate associates with RNA-induced silencing complex (RISC) proteins which degrade one RNA strand
functional RISC formed→ ssmiRNA (22 nucl) and RISC proteins
proteins help miRNA bind 3’UTR
What are the functions of the RNA-induced signaling complex (RISC) proteins? There are two important functions.
target mRNA binding using miRNA w/ complementary base pairing
gene silencing through mRNA degradation or translation inhibition
What happens when a miRNA has a perfect match with its target? What happens when there is a partial match?
perfect match→ mRNA degraded
partial match→ mRNA is not translated (halt)
How is one miRNA able to regulate multiple mRNAs?
partial match allows one miRNA to regulate multiple mRNAs
→ one miRNA regulate expression of 400 mRNAs
What is the function of a processing body (P body) in eukaryotic cells?
→ store mRNAs that are not engaged in translation at the cytoplasm
stored and translated later or degraded
What is short interfering RNA (siRNA)?
siRNA: ds RNA that induced degradation using RNAi pathway
Compare and contrast siRNA and miRNA. How are they similar? How are they different?
same?
processed by Dicer in cytoplasm
used as microRNA to reduce gene expression (translational control)
different?
miRNA is transcribed from genome
SiRNA comes from virus (artificially made)
miRNA leads to translational repression or degradation
siRNA only leads to degradation
What is short hairpin RNA (shRNA)? Describe how shRNA is processed in cells.
shRNA: ss of RNA has hairpin secondary structure to form dsRNA
**siRNA either works as a dsRNA or shRNA
used by scientist to knockdown specific genes using viral vectors to insert RNA into genome
once inserted in the nucleus— shRNA can be processed by Drosha in nucleus
Describe how siRNA or shRNA can be used to treat a human disease.
inject siRNA into cells
down regulate the gene
fewer symptoms
Compare and contrast ncRNAs in eukaryotic and prokaryotic cells. What are some ncRNAs that are observed in prokaryotic and eukaryotic cells? What are some ncRNAs that are unique to eukaryotic cells? What are some ncRNAs that are unique to prokaryotic cells?
same? tRNA, rRNA
different?
euk: snRNA, snoRNA, telomerase RNA, lncRNA, miRNA
prok: crRNA & sRNA
What are sRNAs? What is a riboswitch?
sRNA: regulatory RNA (50-500 nucl) that bind mRNA to regulate gene expression
riboswitch: structure in 5’UTR in mRNA that binds to small molecules allowing regulation of gene expression
Describe how sRNAs and riboswitches interact to control gene expression.
sRNA bind to riboswitches to promote (positive regulation) or inhibit (negative regulation) translation
What is the function of the CRISPR-Cas system in bacteria?
→ prok immune system that is used by bacterua to defend against viral infections
The CRISPR Cas system uses two non-coding RNAs and different Cas proteins. Describe how the CRISPR-Cas system works in prokaryotic cells and explain the role of non-coding RNAs and Cas proteins in this process.
adaptation phase: bacteria integrates viral DNA into chromosome
Cas1/Cas2 cut up viral DNA
expression phase: bacteria express crRNA (integrated viral DNA), tracrRNA, and Cas9 proteim
tracrRNA (scaffold) bind crRNAs (guide) using complementary base pairing
interference phase: crRNA detects second viral infection
tracrRNA-crRNA complex binds Cas9 (a nuclease)
crRNA recognized viral DNA and Cas9 destroys it
What is transgenesis?
→ process of transferring a gene (transgene) from one organism to another
**characteristic of organism changes because of insertion of new gene
Describe how transgenesis can be performed with bacterial cells. What are some benefits of using transgenesis with bacterial cells?
bacteria transformation: create a recombinant plasmid with bacterial plasmid and studied gene, allow transformation by into bacteria and select for successful cells.
benefits?
make large amounts of human proteins like insulin and human growth hormone
bioremediation: biological organisms used to remove pollution from air, water, or soil
Identify two types of eukaryotic cells that naturally possess plasmids.
yeast cells
some plant cells
Describe the 2-micron plasmid. What type of cells naturally possess the 2-micron plasmid? What were some important sequences that were discovered on the 2-micron plasmid? Why were these sequences important?
discovered in yeast nuclei
sequences?
yeast origin of replication (Ori)→ allow plasmid replication
centromere sequence (STB)→ allows plasmic to participate in mitosis
What is a shuttle vector? What are the functions of the following sequences in a shuttle vector: antibiotic resistance gene, recognition sites, two different origins of replication, centromere sequence, and selectable marker.
shuttle vectors: plasmids that can propagate in two different host species
sequences?
antibiotic resistance gene: selectable marker in bacteria→ make many copies of recombinant plasmid
recognition sites: DNA sequences recognized by RE, where gene is inserted
two diff origins of rep: one for year and one for bacterial replication
centromere sequence: allow mitosis participation in yeast
selectable marker: allow selection after yeast transformation
What is selective breeding?
→ modify phenotypes in economically important species through non-random mating
**only cross individuals with desirable phenotype over many generations
What is inbreeding depression? What is the association between selective breeding and inbreeding depression?
inbreeding depression: inbreeding of several generations promotes homozygosity for deleterious alleles
**selective breeding leads to inbreeding depression
Provide some examples of selective breeding that can be observed today.
size of agricultural animals→ increased 4X
domestic animals→ dogs
variety of agricultural plants
What is a genetically modified organism (GMO)?
→ organism that has its genome modified by laboratory techniques
What is the difference between a GMO and an organism created through selective breeding? What is a similarity?
same?
enhance desirable traits
artificial— not found in wild
different?
GMO→
directly alter organisms genome in lab
change in one generation
selective breeding→
rely on existing genetic variation
change over many generations
What is a transgenic plant?
→ plants that has been genetically modified to create a new trait
What is a Ti plasmid? What is T DNA? Describe how a Ti plasmid can be used to make a transgenic plant.
plant-infecting bacteria contain Ti plasmid that can transfer T DNA to the plant genome upon infection
transgenic plants? Ti plasmid used as a vector
T DNA vector is transformed into bacteria→ transformed cells have T DNA vector
transformed bacteria infect plant cells→ T DNA is inserted into plant genome
single transformed plant cell used to make entire plant
Describe at least two examples of GMOs that can be observed today. What is beneficial about their genetic modification?
Roundup Ready Crops: plants contain genes that makes them resistant to Roundup herbicide
Bt crops: plants contain gen that makes a toxin which kills a variety of insects
AquAdvantage salmon: salmon contain gene for growth hormone that increases growth
benefits? increase yield
What is gene therapy? How does gene therapy treat a genetic disease?
gene therapy: experimental technique that can introduce cloned genes into human cells to treat or prevent disease
treat genetic disease?
viruses are used to insert cloned genes into human chromosome to treat autosomal recessive diseases
monogenic diseases→ deliver one cloned gene to address disease from non-functional gene
aa→ Aaa
What is a viral vector? Describe the process of gene therapy in three steps.
viral vector: modified viruses (cannot reproduce) that deliver genes into human cells to restore function of cells causing disease
steps?
gene of interest is cloned into viral genome
recombinant virus (w/ gene of interest) infects human cells that are causing disease
virus uses natural ability to insert viral genome into human chromosome (gene addition)→ gene of interest inserted in human chromosome and cell has one functional gene to cure disease
Describe how LUXTURNA is used to treat blindness.
cause of disease? LOF mutation at gene called RPE65
surgeon injects viral vector to cells of eye that cause blindness
virus deliver functional copy of RPE65 gene to restore function
gene present on circular episome (extrachromosomal DNA)
What is gene editing? How is gene editing different from gene therapy? How is it similar?
gene editing: describes process of making targeted modifications to endogenous (pre-existing) gene in living cell
same? cloned gene inserted into organism
different? gene editing is targeted while gene therapy is random insertion
How was gene editing performed in the 1980s? What was the problem with gene editing in the 1980s? What was the solution to this problem?
1980s? embryonic stem cells could use homologous recombination to insert cloned gene into mammalian chromosome
issue? low efficiency
solution? use target nuclease (CRISPR/Cas 9 system) to cut DNA at specific location
Why is the CRISPR-Cas9 gene editing superior to the methodology used in the 1980s?
CRISPR-Cas9 system is used to target a nuclease to a specific location and promote gene editing
how? two components→ a guide (gRNA) and nuclease (Cas9)
Why is a double-strand break (DSB) important for gene editing?
DSB triggers the cells natural repair mechanism→ naturally initiate recombination of gene of interest into chromosome
Summarize how the CRISPR-Cas9 gene editing system works using the terms gRNA, Cas9, donor DNA, homologous recombination, and nonhomologous end joining. Try to describe the function of each term in the summary.
program CRIPSR mechanism to bind specific sequence using guide (gRNA)
harness nuclease of mechanism (Cas9) to make a targeted cut of DNA
repair the DNA:
donor DNA present (copy present): homologous recombination→ fix DSB and gene replacement occurs
donor DNA absent (cope absent): non-homologous end joining→ fix DSB and deletion mutation occurs
The CRISPR-Cas9 system can be used to make different types of genetic modifications. Describe these genetic modifications and explain how scientists can choose which type of genetic modification will be generated.
HR or NHEJ→ scientist can either provide or not provide a copy of the donor DNA
yes copy→ HR→ no deletion mutation
no copy→ NHEJ→ yes deletion mutation
What is the main cause of sickle cell disease in human patients?
DNA mutation→ abnormally functioning red blood cells which are unable to transport oxygen throughout organism efficiently
Describe some differences between fetal hemoglobin and adult hemoglobin. When are they used during human development? What subunits are in fetal hemoglobin and adult hemoglobin?
hemoglobins:
fetal→ alpha and gamma subunits (used from fertilization to 30 weeks old)
adult→ alpha and beta subunits (used from 30 weeks to death)
Describe the relationship between sickle cell disease and adult hemoglobin.
SCD patients have mutation in beta subunits of hemoglobin so disease→ symptoms begin after transition to adult
Describe how Casgevy is used to treat sickle cell disease. In your description use the following terms: blood stem cells, BCL11A, GATA1, adult hemoglobin, beta hemoglobin, gamma hemoglobin, and fetal hemoglobin.
Treatment?
collect blood stem cells from patient w/ SCD
use CRIPSR to modify blood stem cells so normal hemoglobin is produced
transfer modified blood stem cells back into patient
blood stem cells produce normal blood cells→ SCD symptoms decreased
Mechanism?
use CRIPSR to modify enhancer of BCL11A→ a RTF that inhibits gamma subunit expression in adults
NHEJ preformed and RTF no longer produced
gamma subunit gene is expressed→ adult RBC have alpha and gamma subunits
What is DNA sequencing (aka Sanger sequencing)?
→ determining the sequence of bases for a fragment of DNA
sequencing=replication in a tube
What are the critical components of a DNA sequencing reaction? What is the function of each component?
components:
template strand of DNA: desired sequence
one sequencing primer: short DNA strand that is complementary to template and primes for SNA synthesis (provides 3’OH)
nucleotides (dNTP and ddNTP): add to new DNA strand
DNA polymerase: connect nucleotides to make a new DNA strand
Compare and contrast a DNA sequencing reaction and the polymerase chain reaction (PCR). What are the similarities? What are the differences?
same?
replication in a tube
require primers
require template strand
different?
PCR needs two primers, sequencing needs one
purpose: amplification vs. sequence
products: all same vs varying length
nucleotides: only dNTP vs. both dNTP & ddNTP
What is the function of a dNTPs in a sequencing reaction? What is the function of a ddNTP in a sequencing reaction?
dNTP: used to grow DNA strand in 5’ to 3’ direction bc has a 3’OH
ddNTP: used to terminate DNA synthesis bc lacks 3’OH (has florescent tag)
[dNTP]>[ddNTP]
Describe how the critical components described above are used together in a DNA sequencing reaction using 3-4 sentences.
isolate template strand
use primer, DNA polymerase, and the nucleotides to synthesize strands of varying lengths
sort segments by size in gel electrophoresis
use detector to read fluorescent labels→ computer generates sequence of bases in 5’ to 3’ direction (match color to base)
What is the difference between a gene and a genome?
gene: only DNA sequences that produce a product (RNA or protein)
genome: all genes and non-coding DNA in an organism: all DNA sequences on all chromosomes
What are two challenges to sequencing the genome?
organization: chromosomes are tangled together in nucleus (ball of yarn)
size: human genome has ~25,000 genes and ~3 billion bases, sanger sequencing can only do ~750 bases at a time
What is the human genome project? How long did it take? How much did it cost?
human genome project: international collaboration to sequence entire human genome using DNA sequencing technology
13 years
$3 billion dollars
What is the reference sequence? What is the association between the human genome project and the reference sequence?
reference sequence: high quality genome sequence, product of the Human Genome Project
What were some of the major issues with the human genome project?
took too long
was too expensive
why→ use of gel during DNA sequencing
What is next generation sequencing (NGS)? How does NGS address the issues encountered during the human genome project?
NGS: sequence millions of DNA fragments simultaneously without using a gel
Describe the process of NGS in 3-4 sentences. In your description explain the function of the adaptor sequence, sequencing primer, and reversible terminators.
isolate genomic DNA and cut into small fragments; adaptor sequences attached to both ends of fragments
attach small fragments to chip (~200 million fragment into 200 million well)→ use adaptor to attach to each well
use reversible terminator sequencing to sequence 200 million frags simultaneously; DNA is sequenced as new strand is being made (no gel)
sequencing primer binds to adaptor sequence
camera takes pictures after each round of halting-nucleotide exposure, reversal to synthesizing-nucleotide, and add next round of halting-nucleotides
computer assembles genome sequence using sequence from DNA fragments and reference sequence
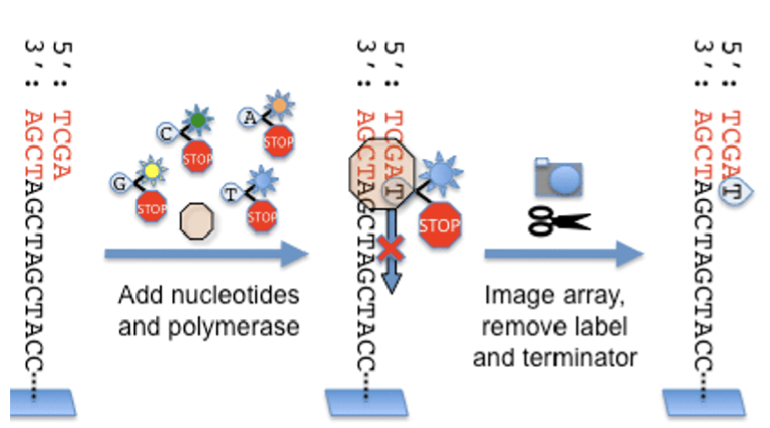
Compare and contrast ddNTPs and reversible terminators. How are they similar? How are they different?
same? halt synthesis, emit fluorescence
different? reversible terminators are have a removable fluorescence and 3’ blocking group
Explain the relationship between the human genome project and next generation sequencing. How does one depend on the other?
NGS generate millions of reads (200 million short DNA segments)→ computer uses reference sequence (HGP) to align reads
What are some advantages of NGS? What is a disadvantage of NGS?
advantages: cost (much cheaper) & time (much faster)
disadvantages: not as accurate as DNA sequencing
What are single nucleotide polymorphisms (SNPs)? Describe how scientists can use SNPs to link genes to diseases.
SNP: DNA sequence variation at one nucleotide that is common in a population
**genomes of two unrelated individuals are 99.9% identical→ majoirty of 0.1% difference is composed of SNPs
relate to disease:
perform NGS on people with specific disease
perform NGS on people w/out disease
compare sequence to identify SNPs associated with disease
What is pharmacogenomics? How can pharmacogenomics influence how doctors prescribe medication to patients?
pharmogenomics: how genes and SNPs affect a person’s response to a drug
impact of treatment→ personalized medicine: doctors use ^ to separate patients into group that receive drugs that maximize effect for disease
Compare tumor testing and germline testing. What is the difference? When is it appropriate to use tumor testing on a patient? When is it appropriate to germline testing on a patient?
tumor testing: identifies mutation in tumor cells→ useful for personalized medicine (identify drug specific to patient)
germline testing: identifies germline mutations associated with hereditary cancer→ germline mutations are in every somatic cell
same? genetic testing technique
different?
germline: performed on blood/saliva, patients may be unaffected, determine risk of hereditary cancer
tumor: performed on tumor cells, patient has cancer, determine prognosis and identify treatment options
Describe the advantages and disadvantages between single-gene and multi-gene genetic tests?
single mutation: analyze DNA sequence at one gene
advantage: good for germline testing bc of specificity
disadvantage: no detecting other mutations
multiple mutation: analyze DNA sequence at many genes
advantage: good for tumor testing→ analyze more genes to increase prob of detecting critical cancer mutation & facilitate effective therapies
disadvantage: identifying more mutations means more variants of uncertain significance (VUS)→ begin or pathogenic mutations discovered that can cause anxiety
What is genomic imprinting? Describe the difference between imprinted genes and non-imprinted genes.
genomic imprinting: rare genes→ genes are expressed from ONLY one chromosome in a part-of-origin specific manner
non-imprinted genes: most genes in genome→ genes of two chromosome expressed equally
If imprinted genes are rare, why are they often clustered together?
clustering allows multiple imprinted genes to be regulated by one imprinting control region (ICR): regulatory sequence with a large amount of CpG sites (CpG island)
What is an imprinting control region (ICR)? Describe the function of an ICR.
ICR: regulatory sequence with a large amount of CpG sites (CpG island)
**methylated at one chromosome→ different methylation at ICR allows expression from one chromosome
Are imprinted and non-imprinted genes regulated by ICRs?
only imprinted genes are regulated by ICR
→ ICRs are methylated on one chromosome
Is DNA methylation used to regulate the expression of imprinted and non-imprinted genes? Explain your reasoning.
DNA methylation is used to regulate expression of both types of genes
imprinted: methylation is parent-of-origin-specific
non-imprinted: methylation is general
What is an epimutation? How is an epimutation similar to a DNA mutation? How is it different?
epimutation: change in DNA methylation that causes a change in gene expression→ occur at ICRs to alter expression of imprinted genes and cause developmental defects
same? impact gene expression, heritable, associate with disease, caused by environmental exposures
different? epimutations do not change in DNA sequence and are reversible
Describe how DNA methylation regulates gene expression at the H19/Igf2 imprinting cluster.
H19/Igf2 imprinting cluster regulated by one ICR methylated at paternal chromosome 11
H19: maternally-expressed imprinted gene
Igf2: paternally-expressed imprinted gene
regulates size of tissues/organs
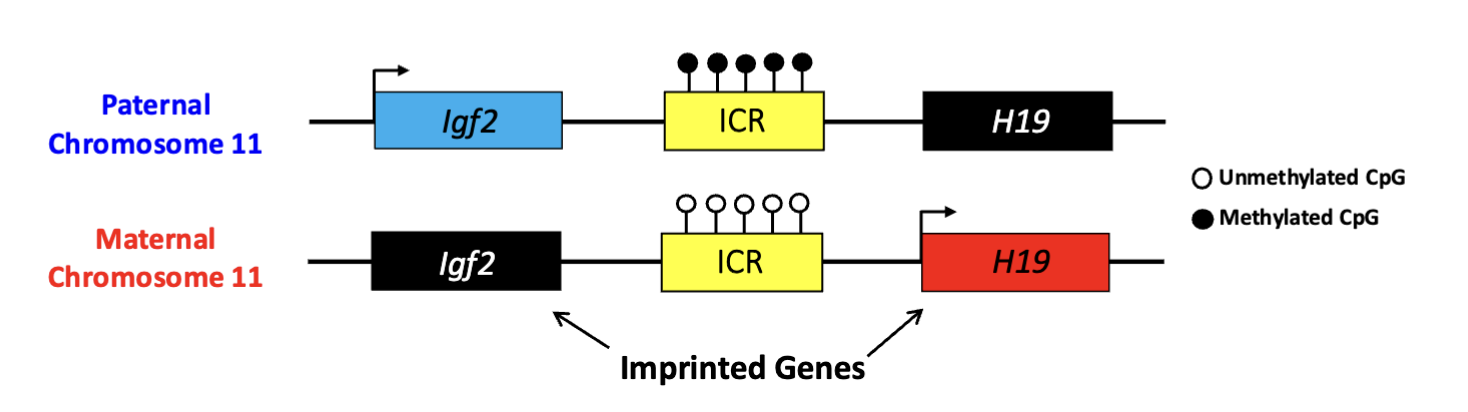
What is a possible of consequence of an epimutation at the H19/Igf2 cluster?
epimutation promotes expression of Igf2 from both chromosome cause BWS in humans (an overgrowth syndrome)
Describe the early stages of mammalian development using the following terms: diploid zygote, somatic cells, germ-line cells, haploid gametes, mitosis.
all diploid organisms use meiosis to create haploid gametes
haploid gametes fuse during fertilization to make diploid zygote
zygote undergoes multiple rounds of mitosis to create early embryo
What is epigenetic reprogramming?
→ genome-wide erasure of epigenetic marks, such as DNA methylation, during different stages of mammalian development
Describe how the H19/igf2 cluster undergoes epigenetic reprogramming in sperm.
DNA methylation is erased in germline cells→ re-established on both chromosomes in gametes (when halpoid sperm is formed all chromosomes are paternal)
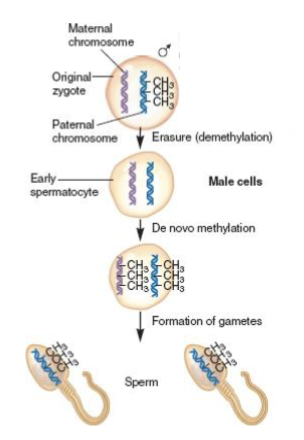
Describe how the H19/igf2 cluster undergoes epigenetic reprogramming in oocytes.
DNA methylation is erased in germline cells→ stays erased on both chromosomes in gametes (when halpoid egg is formed all chromosomes are paternal)
Why is epigenetic reprogramming important for fertilization?
fertilization produces diploid zygote w/ methylated paternal chromosome and unmethylated maternal chromosome at H19/Igf2 ICR
→ established methylation is maintained during mitosis
When does epigenetic reprogramming take place in mammalian development? What two phases?
Describe the epigenetic reprogramming process in both phases of mammalian development.
gametogenesis: production of gametes→ initiated in germline cells of early embryo—completed at onset of puberty
DNA methylation erased at all imprinted and non-imprinted genes during early stages— re-established in late stages
preimplantation: fertilized zygote develops into blastocyst while migrating is fallopian tube—takes ~7 days in human development
DNA methylation erased only at non-imprinted genes—methylation at imprinted genes is maintained
What are assisted reproductive technologies (ARTs)?
→ fertility-related treatments in which eggs, sperm, or embryo are manipulated
most common? IVF
Describe the three main medical procedures that are critical for IVF. What is an IVF cycle?
IVF cycle: combination of three procedures→
superovulation: inject hormones to stimulate ovulation of multiple eggs→ eggs are collected
embryo culture: collected eggs are fertilized with sperm and cultured for 5-7 days in lab
embryo transfer: highest quality embryo(s) transferred into uterus of recipient female
Describe the theory of developmental origins of health and disease.
→ theory that environmental exposures during in utero development influence fetal growth and can predispose an individual to adult metabolic disease
Why was the Dutch famine study able to analyze the developmental origins of health and disease theory?
acute famine in Netherlands during WWII
extreme food rationing/starvation for 8 months due to Nazi embargo
Dutch gov kept accurate pregnancy and birth records during embargo