BIOL 1020: Unit 4 Pt. 1: Cytoskeleton [FINAL]
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
37 Terms
cytoskeleton network main components
3 well-defined filamentous structures that form an elaborate interact + dynamic network
actin filaments (AKA microfilaments), microtubules, intermediate filaments
actin filaments
basic building block of microfilaments; actin proteins polymerize to form actin FILAMENTS
solid, thin structures often organized into a branching network, many of which associated w/ the plasma membrane
most abundant protein in most cells
major contractile protein of muscle cells; major protein in majority of eukaryotic cells
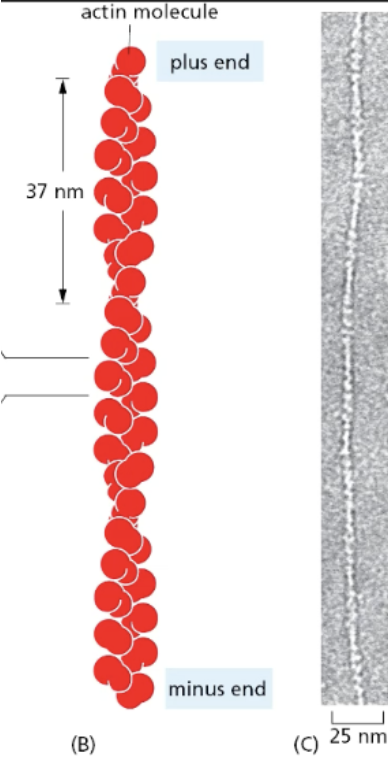
actin in the presence of ATP
polymerizes to form a flexible, helical filament
actin structure
The actin globular subunits (G-actin) in a filament (F-actin) are all oriented in the same direction; they have a (−)/pointed end and a (+)/barbed end. Therefore, actin is polar.
barbed/plus end grows faster than pointed/minus end
growth diffs due to diffs in critical concentration
critical concentrations
the specific concentrations of actin monomers required for polymerization and growth of actin filaments.
require different minimal concentrations of ATP-actin monomers to elongate -> measure of this = CRITICAL CONCENTRATION (CC)
CC of barbed/plus end is much lower than the pointed/minus end = barbed/plus end continues to elongate at lower ATP-actin concentrations than the pointed end can
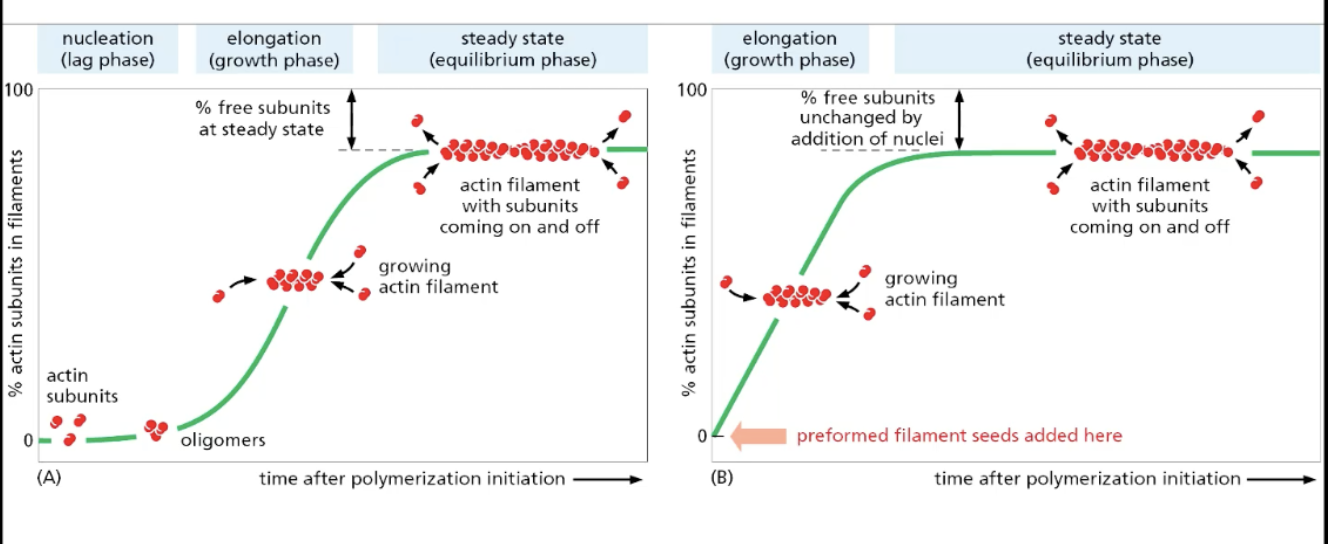
actin polymerization
nucleation (lag phase), in which initial actin subunits come together is energetically unfavorable
bc dissociation rate of actin subunits is higher than the association rate = high chance of falling apart until you have enough actin subunits
once initial actin subunits come together (NUCLEATOR), actin filaments grow at faster rate after bypassing initial nucleation/lag phase
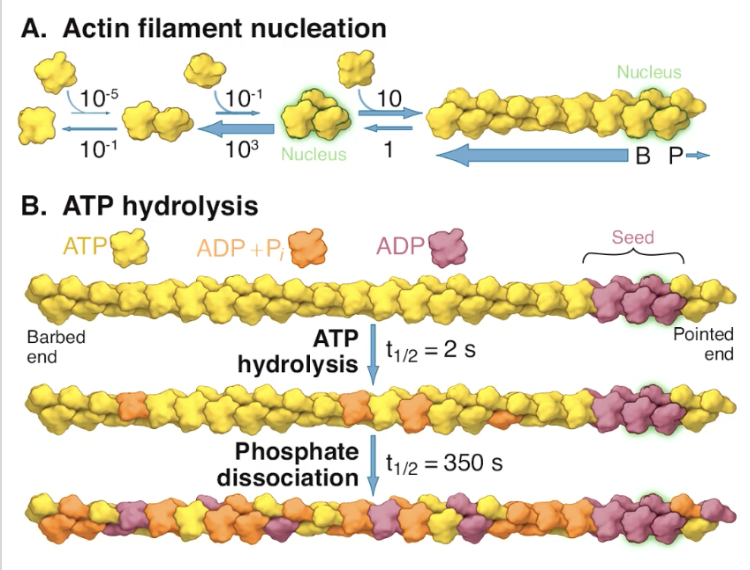
ATP hydrolysis of ATP-actin complex in vitro (how does actin age?)
over time, actin hydrolyzes ATP -> ADP + releases inorganic phosphate = actin aging
more ADP on pointed (minus) end that’s slower growing
differentiates them thru concentration of ADP on either end
critical concentration favors ADP-actin at minus-end, rather than pointed-end
how does actin undergo BRANCHED nucleation?
arp2-arp3 complex (branched actin nucleator)
function
binds laterally to “mother filament” (original actin filament that’s alr there)
acts as nucleator for new actin filaments to form relative to mother filament, which can serve as template for another one using arp ⅔ complex
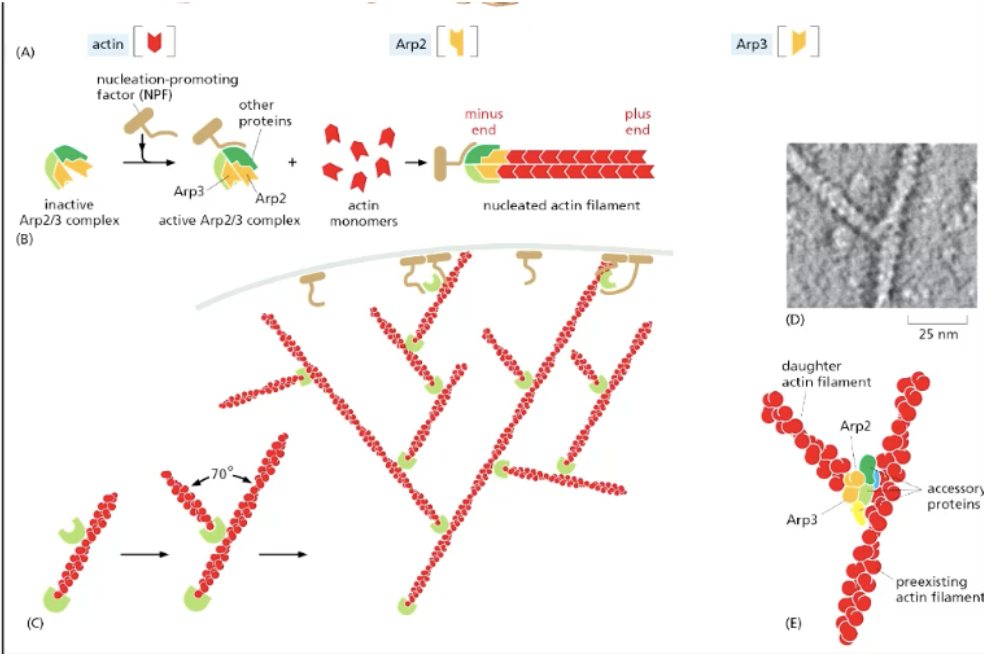
aged actin filaments broken, recycled to create new actin filaments
pointed (minus) end of actin binds to arp ⅔, allowing the barbed (plus) to grow faster outwards to extend the actin branch
how does actin undergo NON-BRANCHED/STRAIGHT actin nucleation?
only polymerize STRAIGHT actin filaments (rather than branches)
how
binds to actin monomers (2) -> creates formin dimer that holds 2 actin monomers tgt = forms nucleus -> formin mediates two actin subnits binding -> actin filament forms
formin moves upwards along plus end as actin subunits aggregate upwards, thus elongating the actin filament
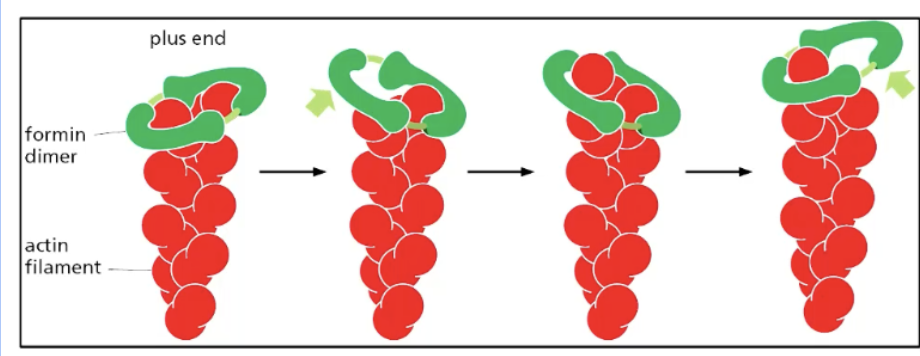
actin function
support cell structure and position organelles properly internally
mediate cell mvts (i.e., cilia, flagella) to propel the cell by exerting force
actin’s role in cellular migration
involves coordinating several proteins to extent the front of the cell (leading edge)
leading edge at front of cell is actin-rich
requires dynamic actin capable of rapid assembly, disassembly, and re-assembly facilitated by actin-binding proteins
myosins
directional motors that move along actin filaments
have DIRECTION; most walked towards barbed (plus) end
function
actin filament polarity (plus, minus ends) -> recognizes diff orientation of actin subunit in filament -> associates to actin filament directionally
“walks along” + moves actin filaments by hydrolyzing ATP as ther E src
myosin II
form of myosin responsible for muscle contraction
myosin II structure
dimeric protein with two heavy chains and two pairs of light chains, forming a distinct structure with a coiled-coil tail and two globular heads (N-terminus) that interact with actin filaments
other end is C-terminus
each myosin horizontally antiparallel to each other in clusters = creates myosin II bipolar thick filament in muscle
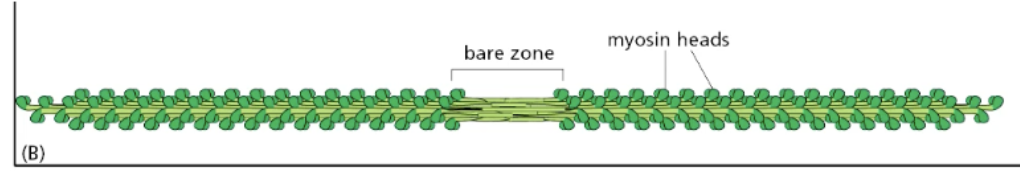
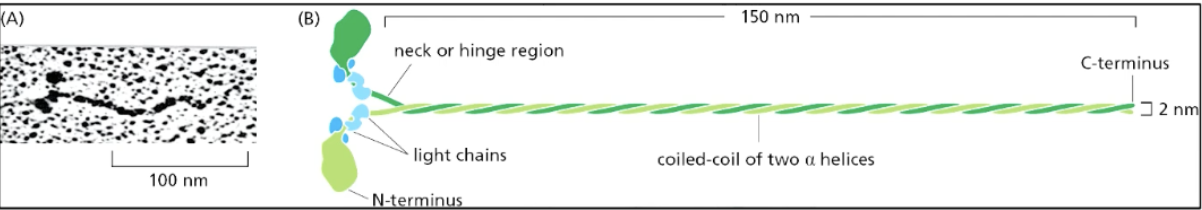
myosin II function in muscle contraction
myosin II sides along actin filaments to drive muscle contraction (in myofibrils of muscle fiber cells)
actin filaments tethered to contractile proteins -> as myosin motors walk along actin -> pulls on contractile protein domain
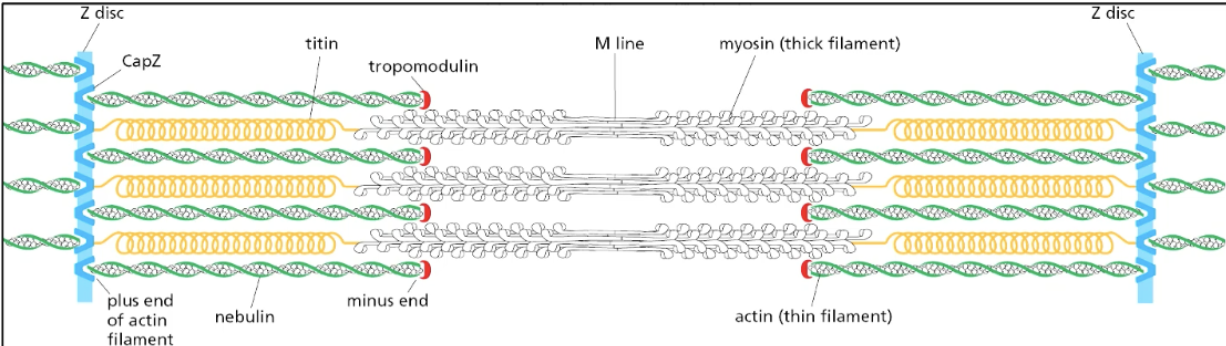
space between Z-disks = 1 contractile unit
motor heads (containining actin-binding domain that binds to actin) walk toward pointed ends of actin on bond sides -> pulls on Z disk structures on both sides -> contracts muscle
motor hydrolyzes ATP -> conformational change -> power stroke where the motor moves to another actin subunit down the actin filament
motor bound to actin is bound v TIGHTLY after one round of contraction
ATP needed to dissociate so that contraction cycle can happen again (i.e., rigor/hardened muscle after death)
elastic proteins extend when pulled + attached to Z disk
What is the structural organization of a skeletal muscle fiber, from cell-level features to sarcomere banding?
Cell-level structure
Skeletal muscle cells are unusually large, cylindrical, and multinucleated
~10–100 µm thick and >100 mm long
Formed by embryonic fusion of many mononucleate myoblasts
Called muscle fibers due to size and multinucleation
Among the most orderly structured cells in the body
Fiber → myofibril organization
Each muscle fiber is a cable composed of hundreds of parallel myofibrils
Myofibrils run longitudinally through the cell
Myofibril → sarcomere organization
Myofibrils consist of repeating sarcomeres (contractile units)
Each sarcomere extends from Z line to Z line
Repeating sarcomeres create the striated (banded) appearance
Sarcomere banding pattern (EM-visible)
Caused by partial overlap of thin (actin) and thick (myosin) filaments
I band: light; contains only thin filaments
A band: dark; corresponds to full length of thick filaments
H zone: lighter region at center of A band; contains only thick filaments
Overlap regions (within A band): contain both thin and thick filaments
sliding filament model of muscle contraction
Core Mechanism: Muscles contract by shortening sarcomeres. Myosin heads (cross-bridges) bind to actin and undergo a power stroke, pulling actin filaments toward the sarcomere center. The filaments themselves do not change length; they slide past one another.
Visual Changes (The "Banding" Rules):
A Band: Remains constant (matches length of thick filaments).
I Band & H Zone: Both decrease in width and can disappear.
Z Lines: Move inward toward the A band.
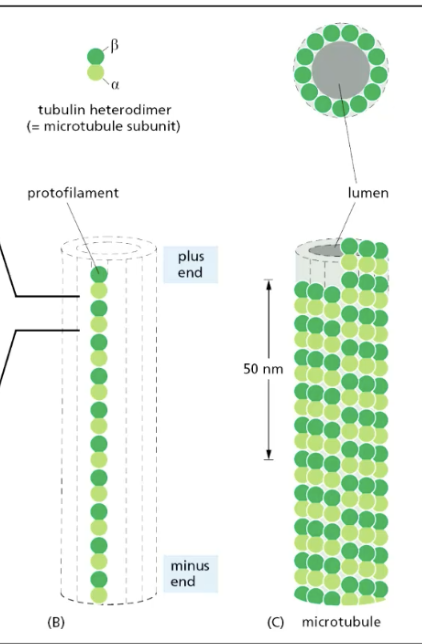
microtubule (MT) structure
rigid = stronger than actin
formed from tubulin heterodimers (alpha + beta) that arrange in head -> tail manner that form PROTOFILAMENT
has plus end + minus end
plus end grows faster
determined by how quickly they hydrolyze GTP -> allows for conformational change -> able to bind to new tubulin heterodimers
12-13 protofilaments arrange in cylindrical structure = microtubule
GTPases
MT heterodimer structure
Free tubulin exists as an αβ-dimer, with the α-subunit binding a trapped and nonhydrolyzable GTP and the β-subunit binding an exchangeable and hydrolyzable GTP. Most of the β-tubulin in microtubules is bound to GDP. In growing microtubules, the (+) ends are capped by GTP-β-tubulin.
plus end = fast growing (row of GTP-beta-tubulin); minus end = slow growing
structural polarity of microtubules is important factor in their growth + function in directed mechanical activities for the cell
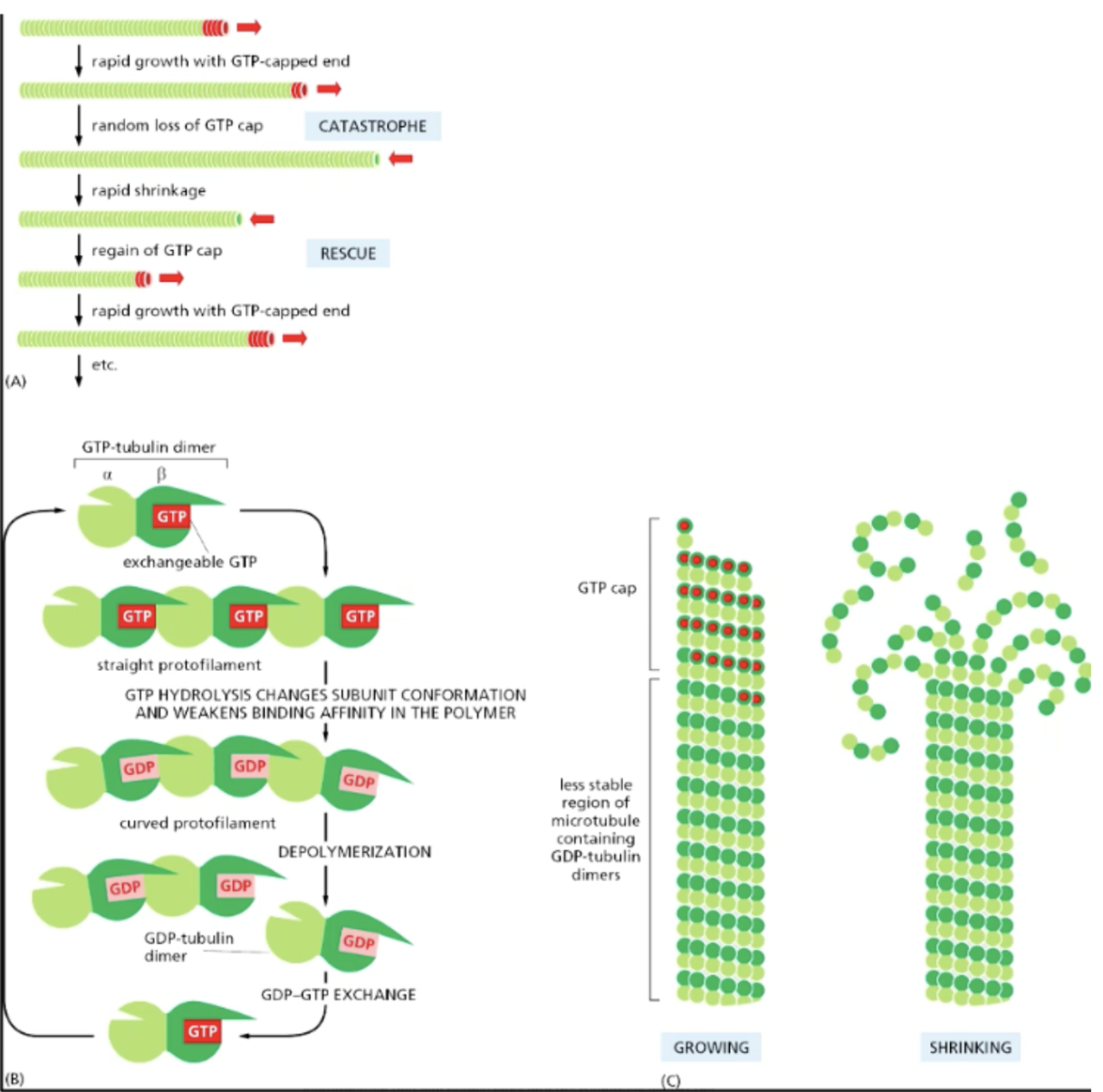
MT dynamic instability
changing between rapid growth + shrinking (catastrophe); happens intrinsically to microtubules (beneficial for cell processes, like division)
growth
happens faster at plus end due to higher rate of GTP hydrolysis
creates GTP-tubulin dimer/GTP-capped end
GTP cap prevents the less stable region of microtubule section containing GDP-tubulin dimers from dissociating/shrinking
during growth periods, tubulin dimers are added faster than their GTP is able to be hydrolyzed → cap of GTP-dimers on MTs at protofilament plus ends, which favors addition of more MT heterodimer subunits
catastrophe
random loss of GTP cap -> rapid shrinkage -> gain of GTP-tubulin dimer cap -> rapid growth
GTP hydrolysis changes subunit conformation + weakens binding affinity in the polymer -> protofilament starts curving -> depolymerization
exchanging GDP w/ GTP “straightens” out the protofilament
purpose of MT dynamic instability
provides mechanism where plus ends of MTs can rapidly explore the cytoplasm for appropriate sites of attachment
allows cells to respond rapidly to changing cell conditions that req remodeling of microtubular cytoskeleton (ex. MT disassembly + remodeling into bipolar mitotic spindle)
how are MTs organized
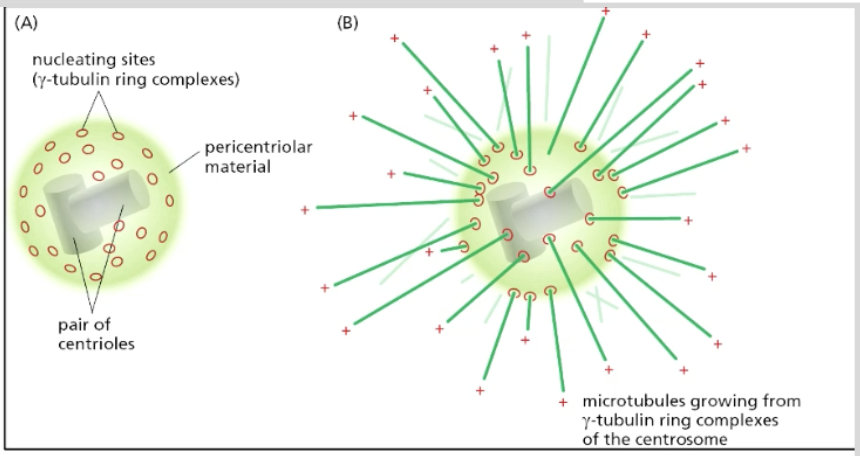
centrosome = main MT-organizing centers (MTOCs) where MTs typically NUCLEATE in animal cells
minus end attached to nucleating sites (gamma-tubulin ring complexes) on the surface of centrosome; plus ends grow outwards (forming a radial array)
radial array usually in undfferentiated cells that contain centrosome near nucleus
cell differentiation -> dissolution/disassociation of centrosome into “bare components” that nucleate MTs -> non-radial form
surrounded by pericentriolar material + has 2 centrioles inside
basal bodies + other MTOCs
centrosomes not the only MTOCs
outer MTs in cilium or flagellum generated direct from MTs in structure called basal bode (resides @ base of cilium or flagellum)
all MTOCs share common protein component =
gamma-tubulin
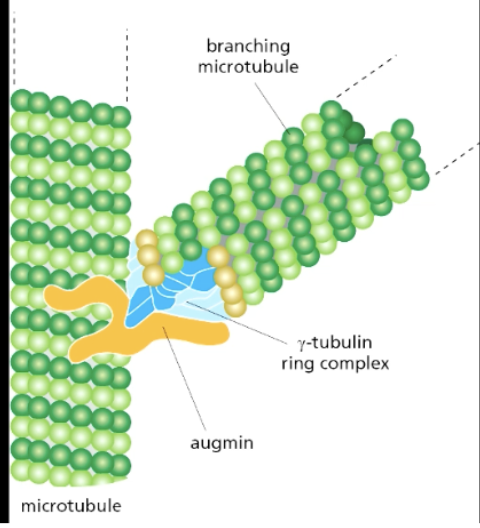
how do MTs branch?
augmin (protein complex) recruits branching MT to nucleate off of existing MT thru gamma-tubulin ring complex
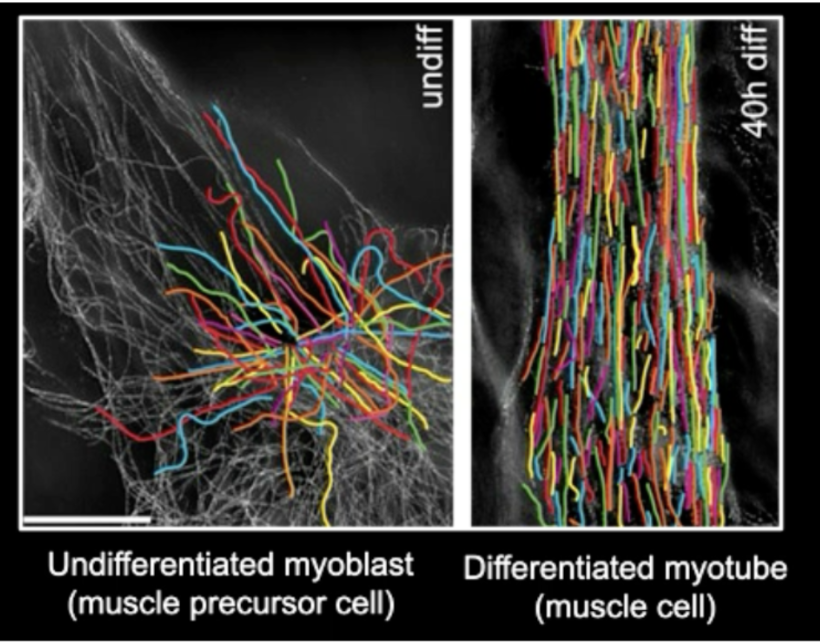
how can MT organization change according to the cell?
diffs along cell types after differentiation (which lends MT organization to the cell’s function)
undifferentiated cells usually have a radial array of MT where they come out from centrosomes, whereas differentiated cells have much more organized MT networks
types of MT-associated proteins
AKA MAPs
+TIPs
structural MAP proteins
microtubule associated proteins
+TIPs
proteins that recognize + associates w/ growing plus ends of MT to connect them w/ other structures (i.e., membranes)
moves along w/ MT as they grow
localizes at plus end “tip” of MTs to recognize the end of MT; continuously associates to MT as long as it grows
doesn't associate w/ MTs that aren’t growing
can track +TIP proteins using GFP to see only the MT plus end tips that are actually growing
structural MAP proteins
crosslink MTs; by linking 1 to another MT
can also associate w/ itself + act as the structure that brings 2 MTs tgt
MAP4 and MAP2
MAP4
crosslinks + creates gap/spacing between MTs most optimally for cell function
more MAPs = more crosslinking/connection between MTs
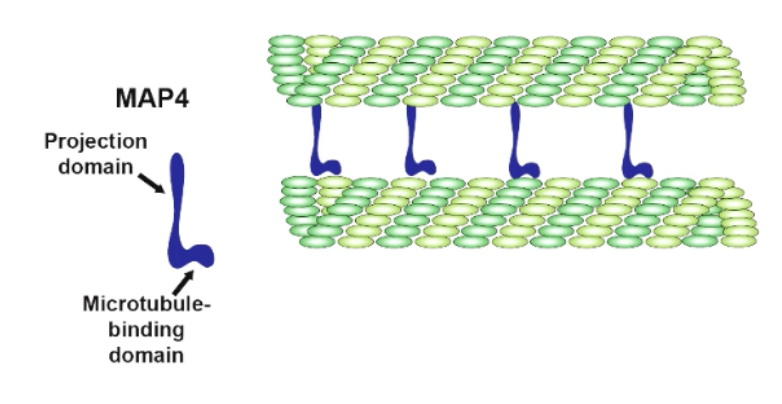
in absence of MAP4, MT gets bunched tgt
zippering of anti-parallel microtubules is important for MT organization in skeletal muscle cells
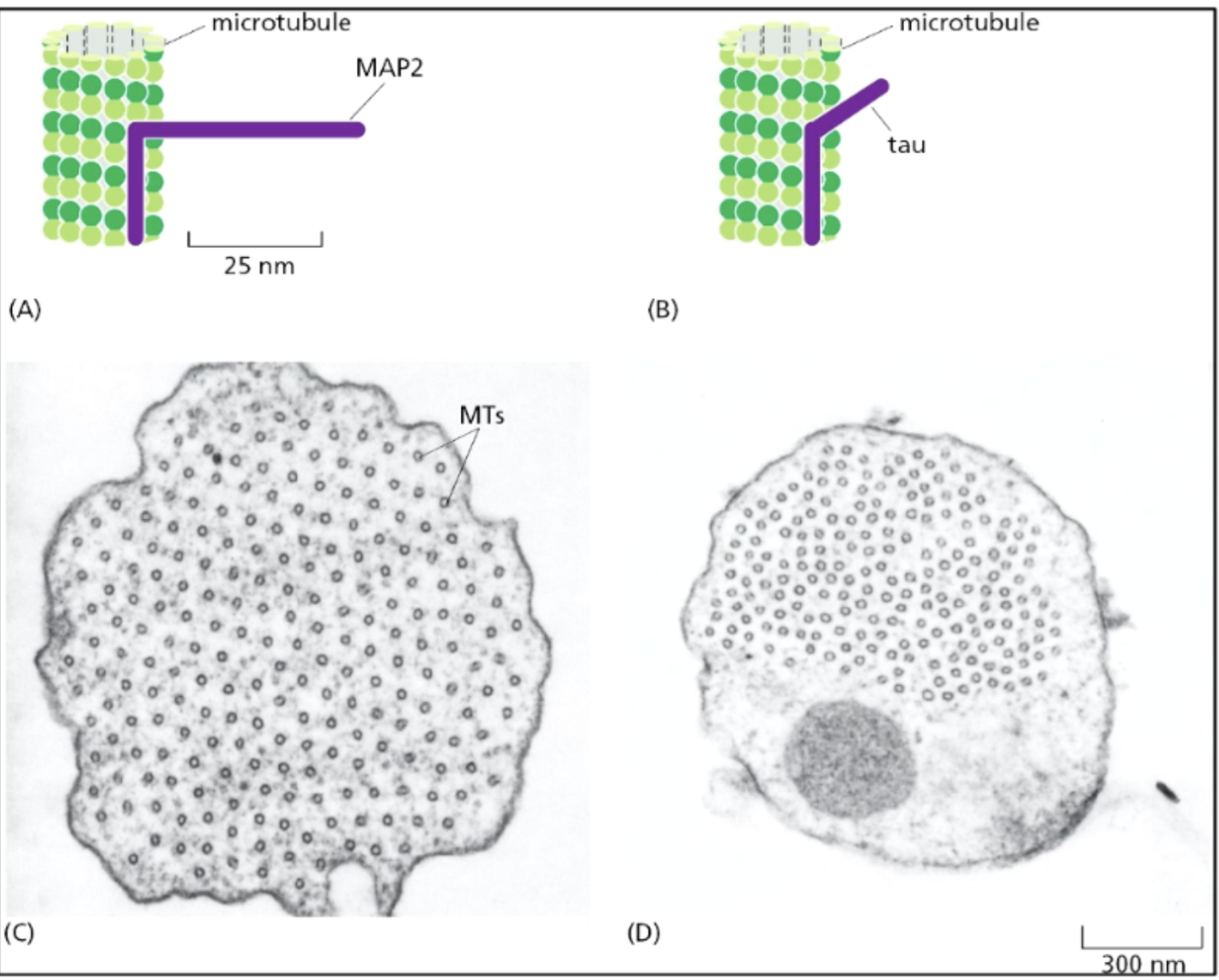
MAP2/Tau family of structural MAPs
length of projection domains of MAPs between MTs in specific cell times determines the spacing of the MTs in the cell + thus cell structure/cytoskeleton
MT motors
kinesins
dynein
kinesin
motor protein that walks along MTs
motor usually walks PLUS END-DIRECTED (N-termius), very few minus end-directed
motor in the middle doesn’t walk; depolarizes MT + controls MT growth
MT depolymerase, where it favors GDP-binding/catastrophe
kinesin 1 structure
dimer
2 head motor domains w/ coiled-coin tail domain connected to light chains
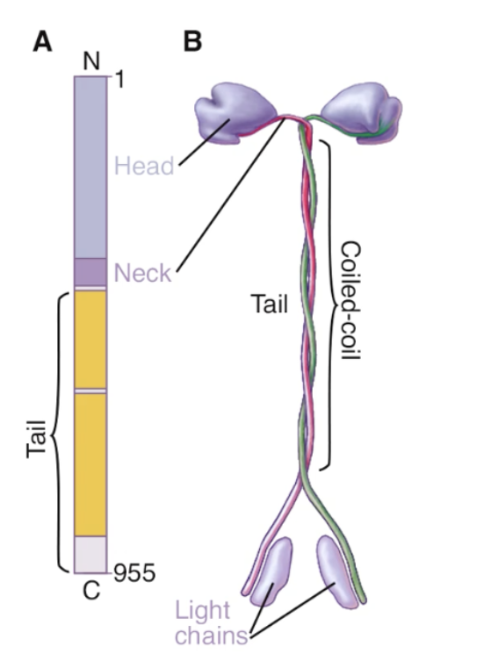
kinesin 1 function
moves along MT thru ATP hydrolysis; ATP hydrolyis undocks kinesin, allowing it to take another step
most processive motors = walks along MT w/o diassociating for long period of time; bc at least 1 motor domain is always held on the MT (as opposed to only having 1 motor domain that undocks often)
often holds vesicle as it moves
dynein
super fast
is a monomer, so can’t innately walk along MT by itself processively
most associate w/ another dynein monomer to move
minus end-directed MT motor
cytoplasmic dynein
encoded by a single gene in almost all eukaryotes but absent in flowering plants + some algae