BIOL 2056 - Mitochondria and Bioenergetics
1/37
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
38 Terms
how do cells get their energy
sugar
amino acids
fatty acids
sunlight
methane
plants generate their energy in chloroplasts
prokaryotes use bacterial membrane
the mitochondria
became encapsuled from another organism
have their own transcription and translation
have a 16Kd genome which encodes rRNA, tRNA etc
requires a large SA for the electrical gradient
mitochondrial matrix
supports the TCA cycle, beta oxidation of fatty acids which is the primary output of NADH
supports the urea cycle, amino acid synthesis and mitochondrial protein synthesis
for these reactions we must get substrates IN
the TCA cycle provides starting materials for:
Amino acids
Porphrins (heams, chloroplasts)
purines and pyrimidines
the mitochondiral membrane
has features which reflects the processes that it supports:
inner membrane is protein rich with a slightly less protein dense outer memb
mitochondrial membrane contains very little cholesterol compared to eukaryotes
high conc of phosphatidylcholine and phosphatidylethanolamine
low conc of Pi and phosphatidylserine
has cardiolipin
4 chains, 2 phosphate groups
anionic lipid
occupies a large volume —> has a small head group and large chains which causes the membrane to curve
outer mitochondrial membrane
must communicate with the mitochondria
has an important role in determining how the mitochondria functions
has porins which make it permeable
mitochondrial porins
beta barrel like structure
relatively unselective
voltage dependent anion channel for the transport of ADP/ATP
the voltage gating comes from the helical chain
lined with positive residues for selectivity
allows ADP/ATP to flow easily
has two conformations: one where the alpha helix is in the middle (closed) and one where the alpha helix moves out the way (open)
interface to cell
it has mechanical links to other organelles and the cytoplasm
regulation of apoptosis —> release of electrochemical gradient regulates cell apoptosis
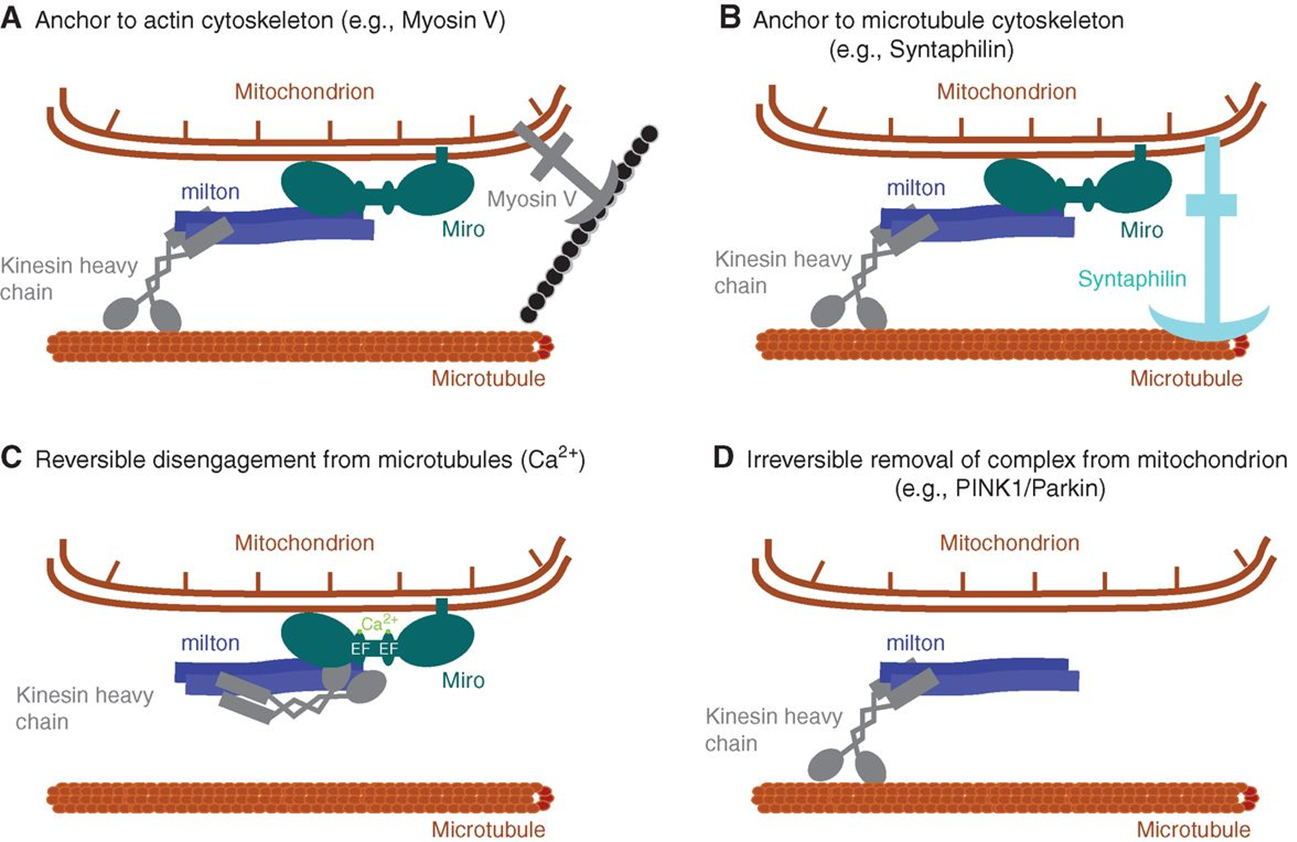
location
mitochondria are distributed throughout the cell
close to the microtubules (colocalised)
the outer mt membrane contains miro which binds to milton —> milton is attached to dynein/kinesin
links to myosin will anchor the mt at a certain place
links to synaptophysin will link the mt to the microtubule
mt can have reversible disengagement from the microtubule motor in a calcium dependent manor
miro can also permanently be broken down leaving a mt at a certain location
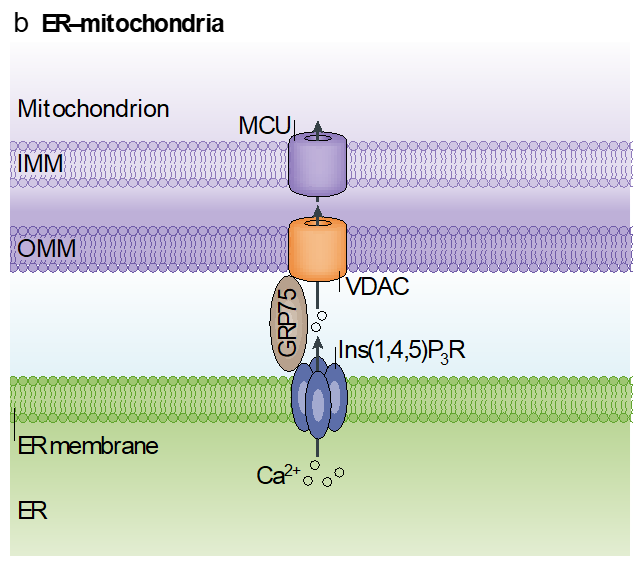
mitochondrial contact sites
mt bound to rough ER
recruit a whole series of molecular components to form contact sites
contact sites facilitate the exchange of lipids
efficient coupling of mt and Er by VDAC adaptors
this supports the metabolic efficiency
energy generation in the inner mitochondrial membrane
transfers high energy electrons from donor to teh high energy terminal receptor
and couples this process with the transfer of protons across the bilayer
F1/F0 ATPase
primary protein for ATP production
F1 head is an enzyme that converts ADP—>ATP
driven by motor complex which is driven by electrochemical gradient
ATP/ADP shuttle
gets substrate into the inner membrane
open face on one side for ADP
conformational change coupled with the binding of ATP
well defined RRRMMM nucleotide binding domain 2×6 transmembrane domains which forms a dimer
shaping membranes
membrane folds round on itself caused by F1 ATPases
fluid mosaic model but although the lipids are fluid, it still has a structure which is controlled by lipids and proteins
MICOS
is a complex that enables tight turning of christae
drives the membrane invagination
composed of MIC10 and MIC60
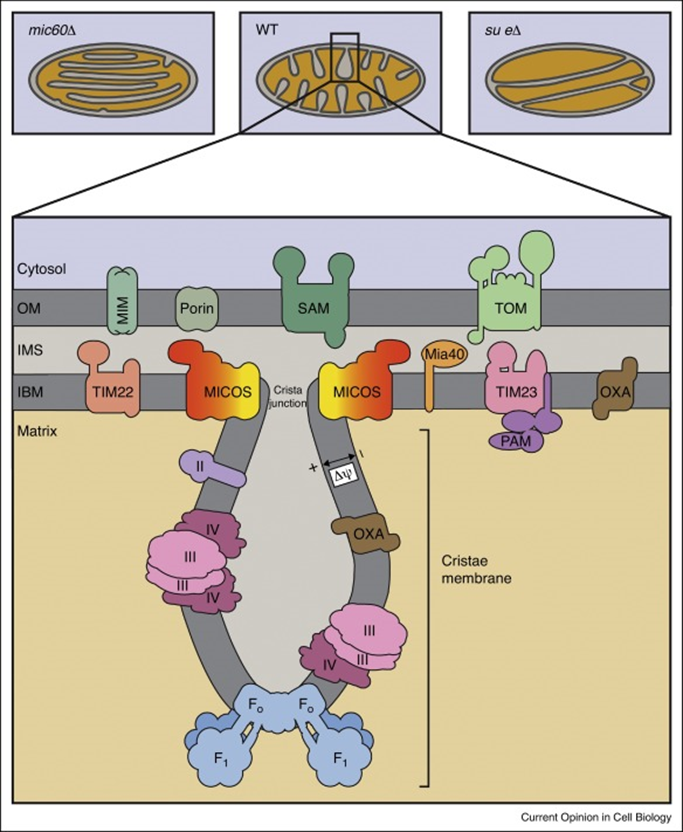
MIC10
main role is cell membrane sculpting
2 transmembrane domains with transmembrane glycine motif and positively charged loop
forms a large oligomeric complex
requires cardiolipin
the glycine disrupts secondary structures and allows efficient packing of the transmembrane domain
the positive loop recruits cardiolipin to the loop
MIC 60
forms a contact between the inner mitochondria and the outer mitochondria
interactions with VDAC, TOM and TIM complex
important to localising activity in the inner and outer membrane
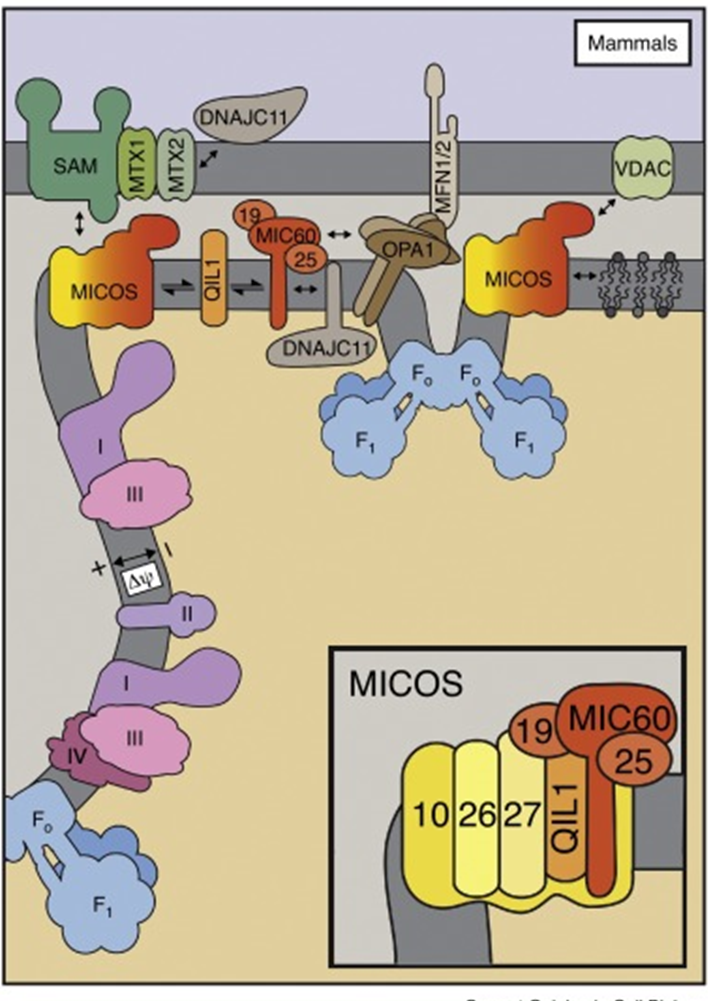
why so elaborate
allows this process to work more efficiently:
advantageous for the electrochemical gradient
ETC is in flatter regions of the cisternae
creates a very small volume which can generate a large change in proton concentration in a very small region
localises the F1 ATPases in close proximity
the electron transport chain
need to be able to capture the energy released
therefore reactions are broken down into smaller steps
there’s no fast, explosive release of energy —> at each step the electrons release energy
the electrons become more positive as they move down the chain
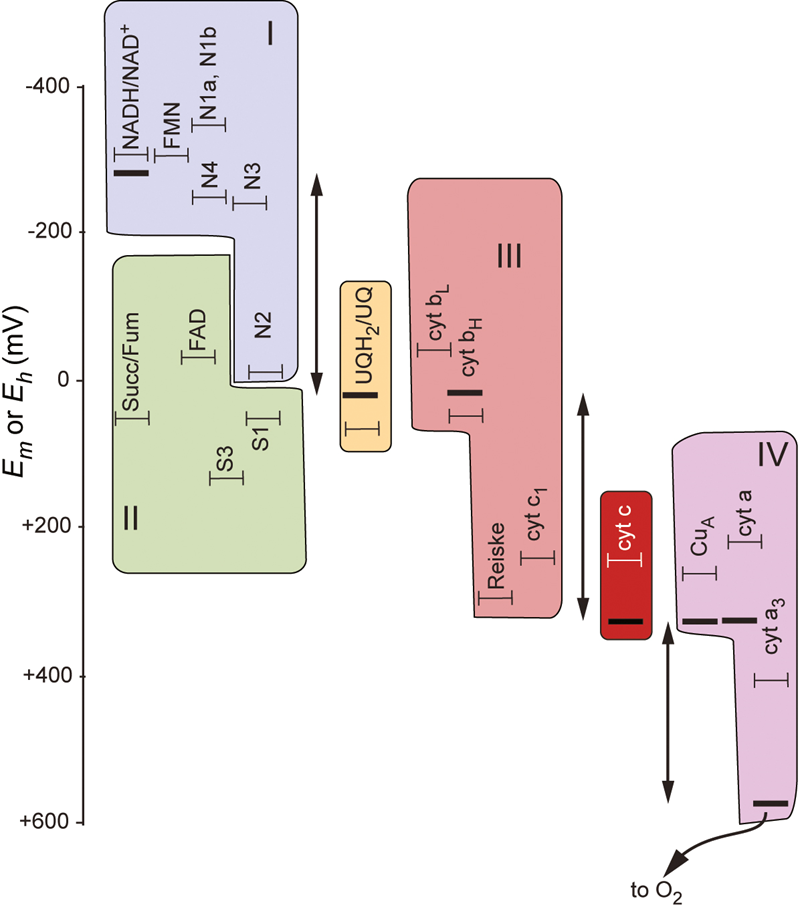
chemiosmotic theory
high energy electrons are used to generate an electron gradient
these high energy electrons then utilise the electrochemical gradient to:
power molecular motors that drive ATP biosynthesis
drive transport of molecules against their concentration gradient
this is conserved in all organisms
the electrochemical gradient
is a gradient of protons —> high concentration outside, low concentration inside
there’s an electrical potential too from the charge
it doesn’t have to be H+
units used are kJ/mol
chemical gradient is deltapH
electrical gradient is deltaψ and represents the change in charge across a membrane
it is often derived in protonmotive force (deltap) which has the units of mV
PREDICTING MOEVEMNT OF ELECTRONS
we can use this equation to derive protonmotive force and predict the moevemnt of electrons where we replace deltap with deltaG
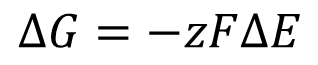
F = the faraday constant
z = charge
Em (denoted the potential at which the pompound is half oxidised and half reduced)
is the tendency of chemical species to acquire electrons
large Em is a high affinity, a low Em is a low affinity
electrons will move towards a larger Em
using the Nernst Equation
using the above equation we can rewrite our equation for free energy for a given mass action ratio
which in terms of electrochemical potential is:
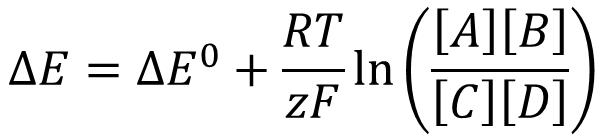
NAD+/NADH REDOC CENTRE
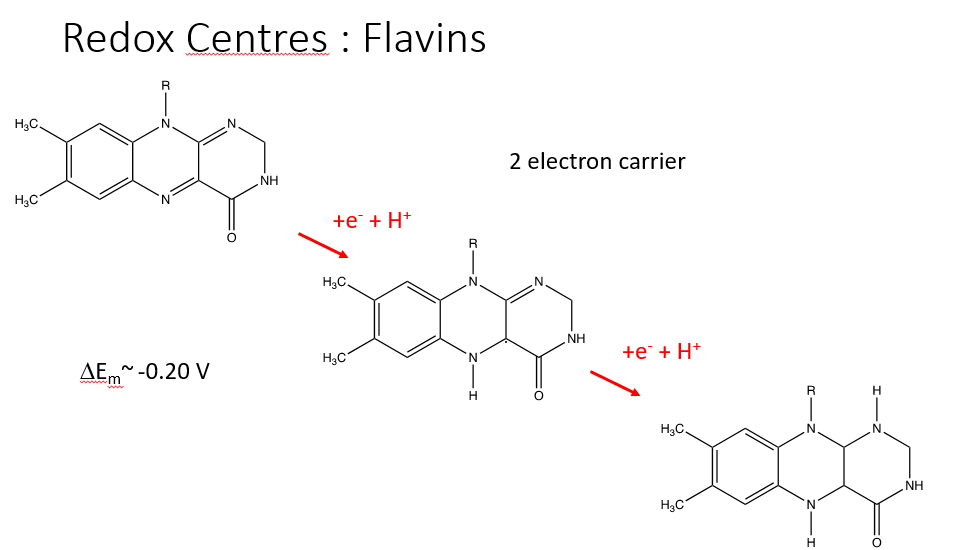
FLAVINS
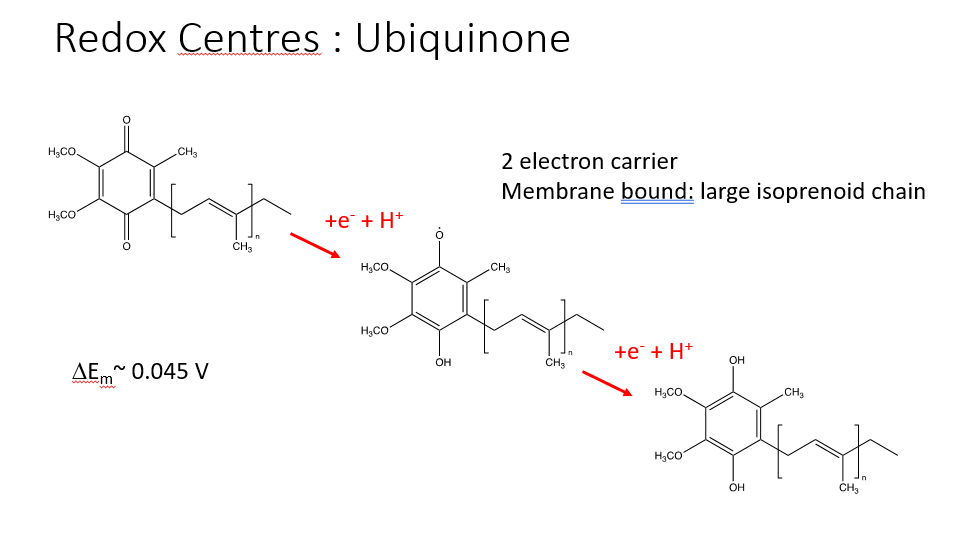
UBIQUINONE
formed in the membrane and is anchored by a large isoprenoid chain
ubiquinone —> ubiquinol
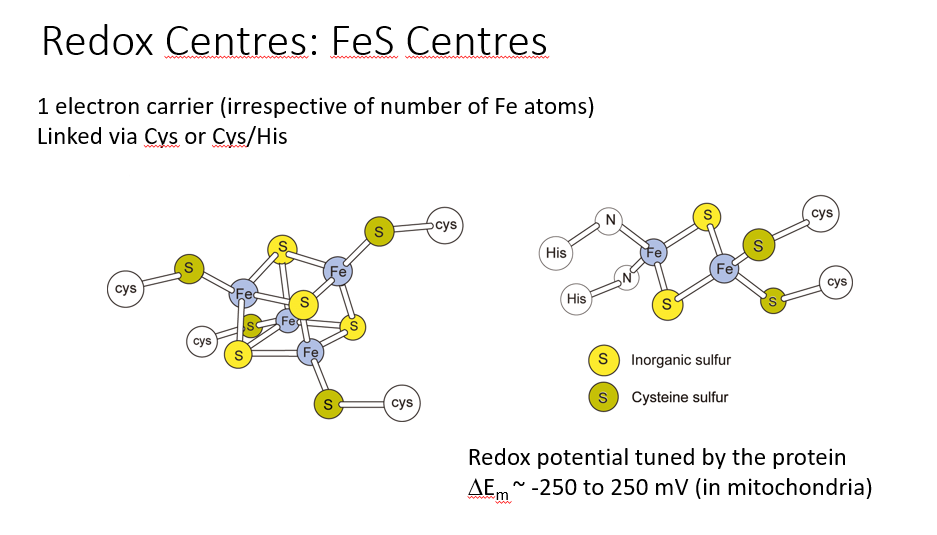
FeS CENTRES
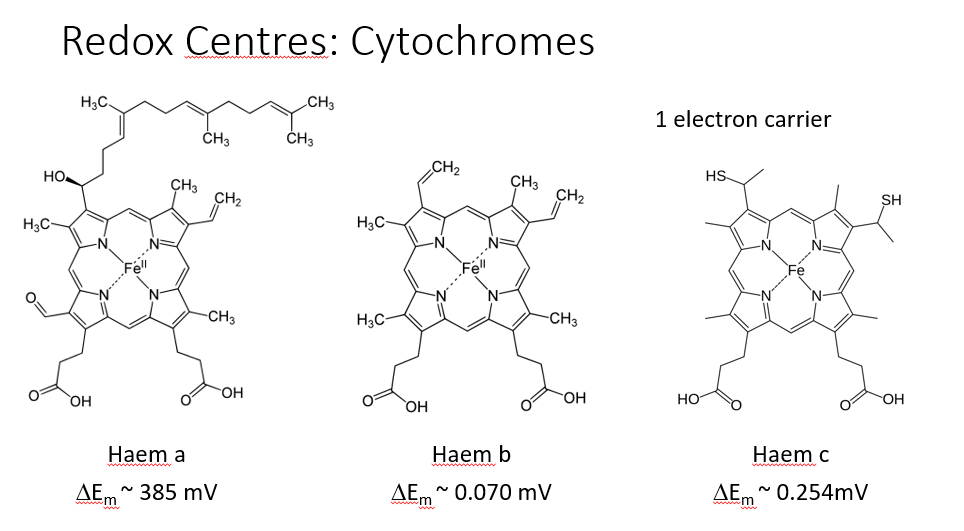
CYTOCHROMES
complex 1 - NADH/Ubiquinone oxidoreductase
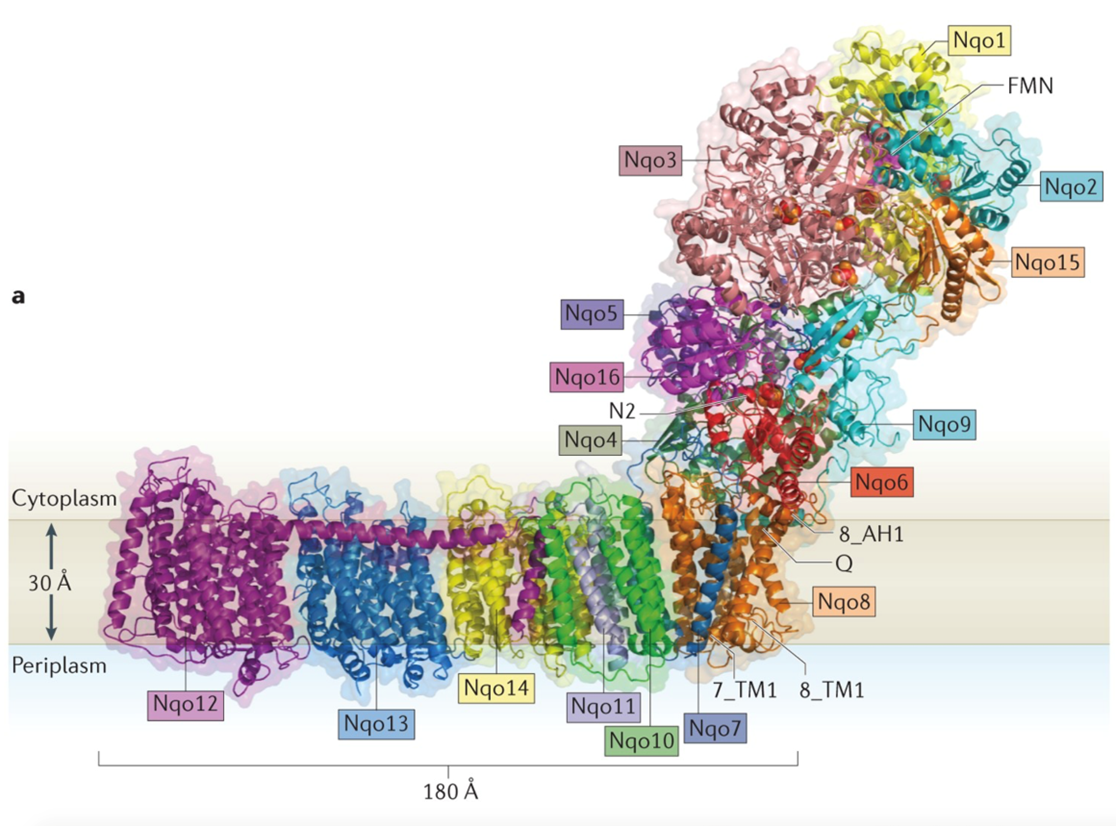
NADH is close to the flavin molecule and the electron jumps to the flavin
it has a transmembrane component and a large extracellular component
A series of FeS centres that work their way down this long arm and the electron uses these to jump down
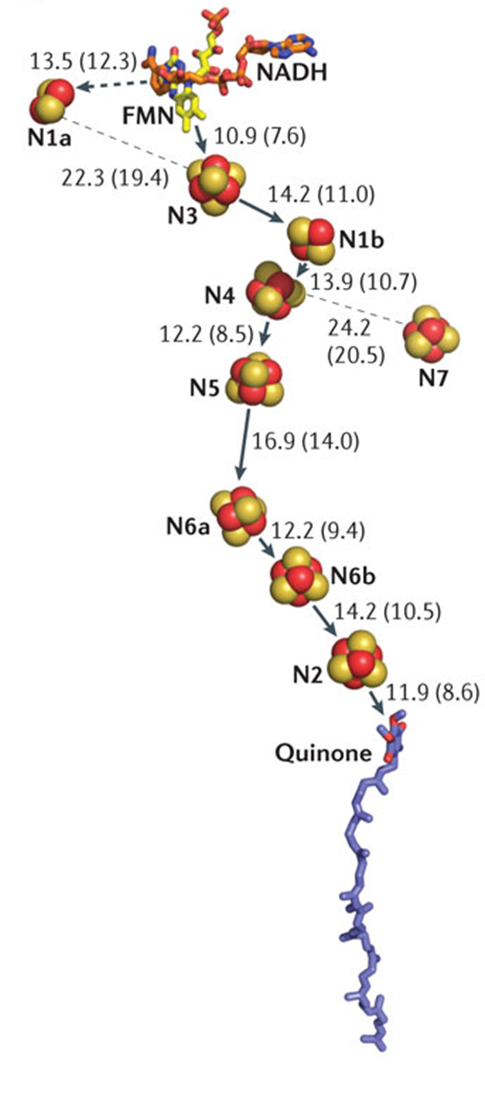
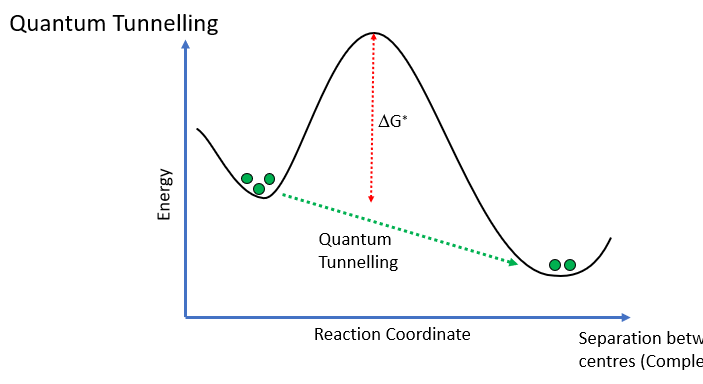
however, they are large distance apart so we would expect the reaction to occur slowly due to large activation energy, however, this doesn’t occur due to electrons quantum tunnelling
moving electrons to the UQ binding site
LOCATION OF THE UQ BINDING SITE
its a 6 membered ring with an isoprenoid chain
its embedded in the membrane
UQ binding site relatively hydrophilic in nature
the transfer of protons from the Tyr/His as Uq essentially pulled out the bilayer
proton pumping of complex 1
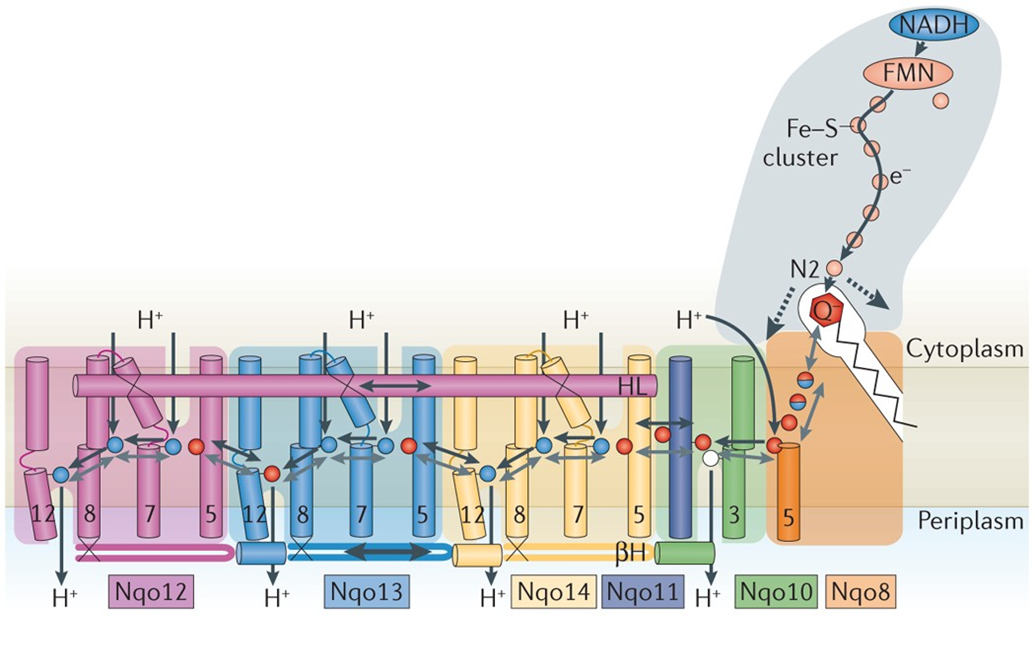
NADH ubiquinone oxidoreductase
there are 5 diffrenet complexes with an alpha helix running paralell through the membrane
the transverse helix reaches out across the pumps to the Uq binding site
the transverse helix couples the conformational change to the energy transfer
it moves back and forth and couples the conformational change to the energy transfer
4 different transport proteins allow the movement of 4 protons across the membrane
complex 3
is a ubiquinone/cytochrome c oxidoreductase
it oxidises ubiquinone and moves the electrons to cytochrome c
ubiquinone carries 2 electrons and cytochrome c carries 1 electron therefore 1 electron must be stored
complex 3 has heam groups
Q cycle
ubiquinone binds to a binding site
reduction of ubiquinone releases 2 protons onto the p face of the membrane
1 electron will move to the FeS centre where it is loaded onto Cyt C
we now have a free radical and so we need to find a way of storing the electron on the ubiquinone
SOLUTION:
the electron is shuttled to cytochrome bL to cytochrome bH
this electron is used to reduce the ubiquinol to ubiquinone
however, we only have one electron added to UQ so this needs to happen twice so UQ—> UQ- —> UQH2
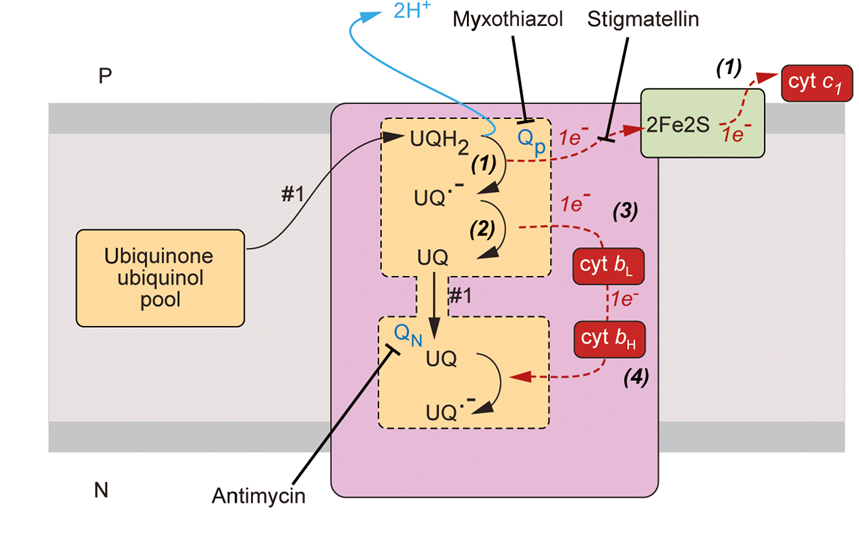
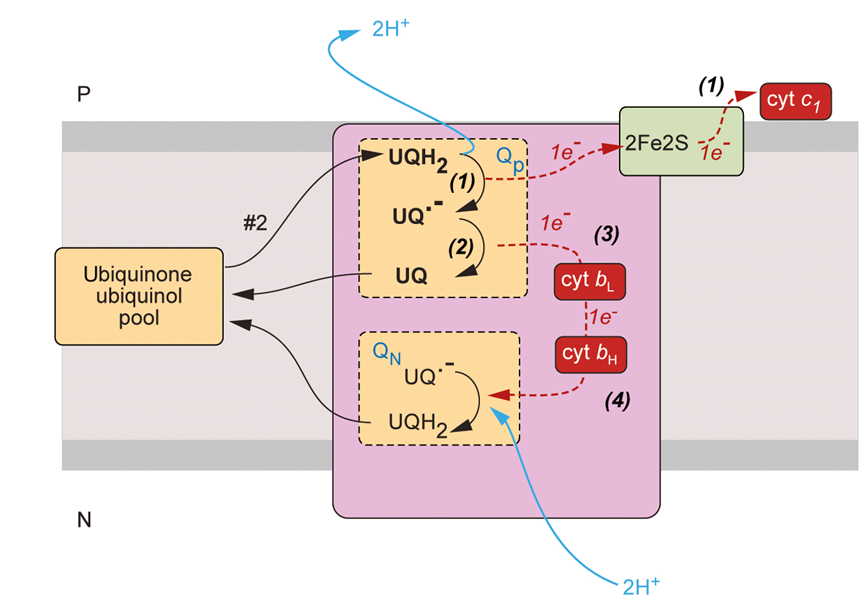
why do electrons take different paths?
repositioning of Rieske proteins directs electrons to different redox centres
cytochrome C
a small molecule with one cytochrome in the middle
one face has a cavity which is a binding site for electrons
has a very positive charge
cardiolipin has a very negative charge so there’s an electrostatic attraction between cyt C and the lipid bilayer
complex 4: cytochrome C reductase
cytochrome C oxidase enzyme
reduce cytochrome C to pass electrons down to molecular )2
adds 4e- to split O2 into water
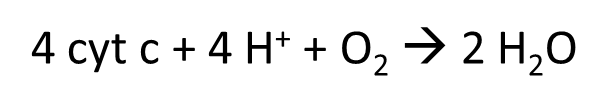
the binding site is at the top which passes electrons down to copper centres to haem centres
couples the loss of energy to the movement of protons
4 protons are pumped out, 4 are reacted with molecular oxygen to make water
has a special haem A3 which also has a copper centre
reduction of O2
occurs at a binuclear centre (cytA3/CuB)
O2 binds to cytA3 which splits O22 and forms a double bond
an additional oxygen is bound to copper B
requires 4e-
2 come from cyt A3 which adopts a 4+ oxidation state, one comes from a tyrosine molecule that becomes a neutral free radical
cytochrome c then delivers an e- and a H+ which restores the tyrosine molecule
delivery of another E- reduces cyt to 3+ state and the H is donated to the O atom which forms a hydroxyl group
further delivery of 2e- and 2H+ restores Cyt A and CuB to their original oxidation state and allows 2 molecules of water to form and dissociate from the complex
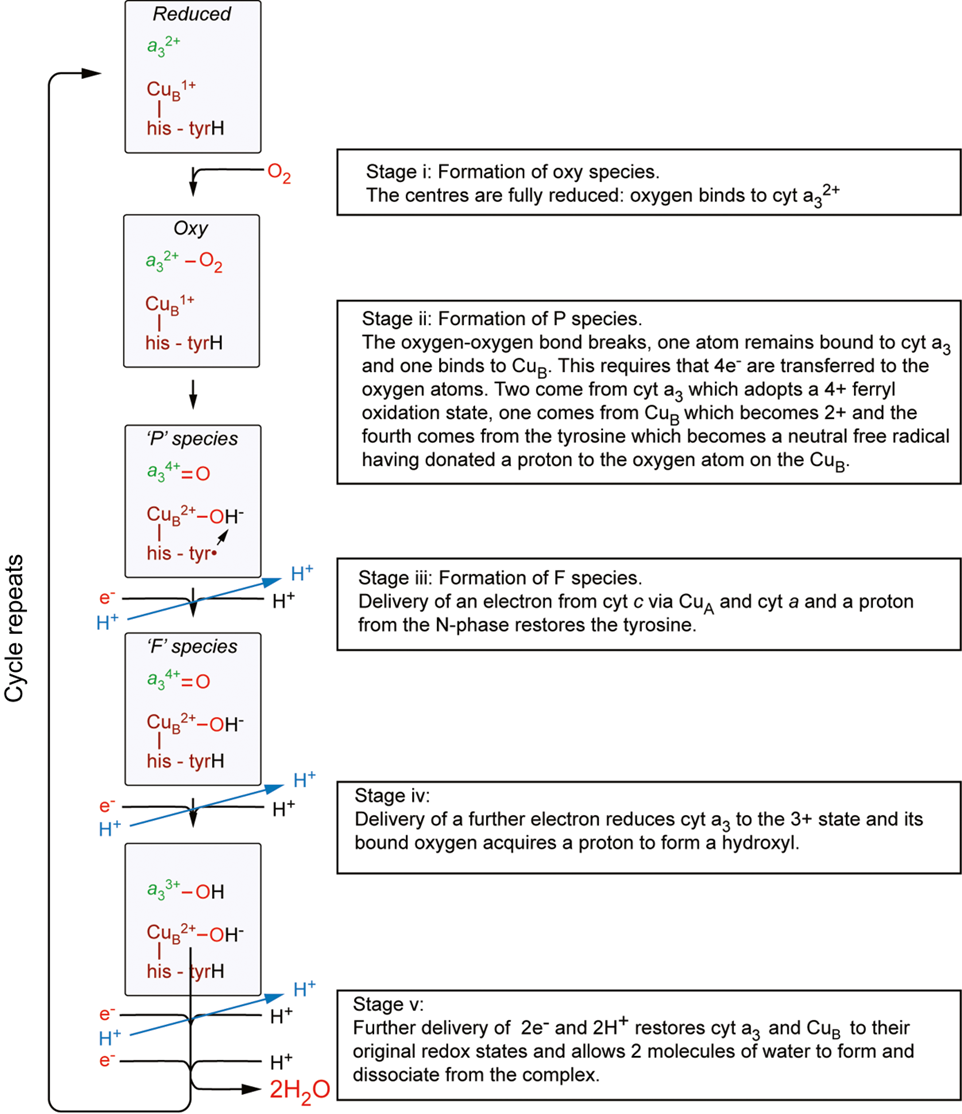
pumping protons
2 channels
D channel: proton pumping pathway
K channel: protons for H2O
there’s a network of charge residues where you can see these protons hopping from one side to another
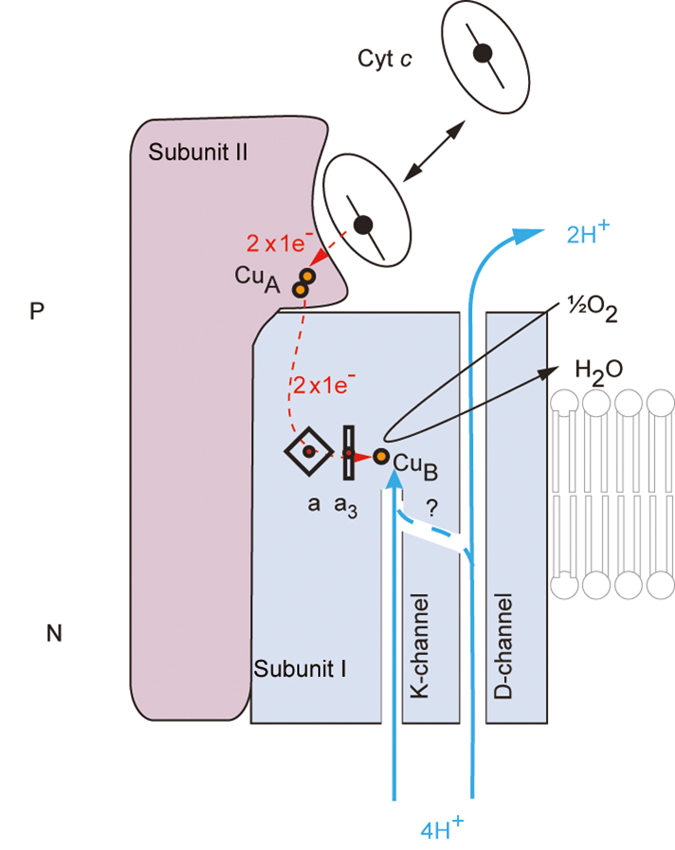
summary: ETC
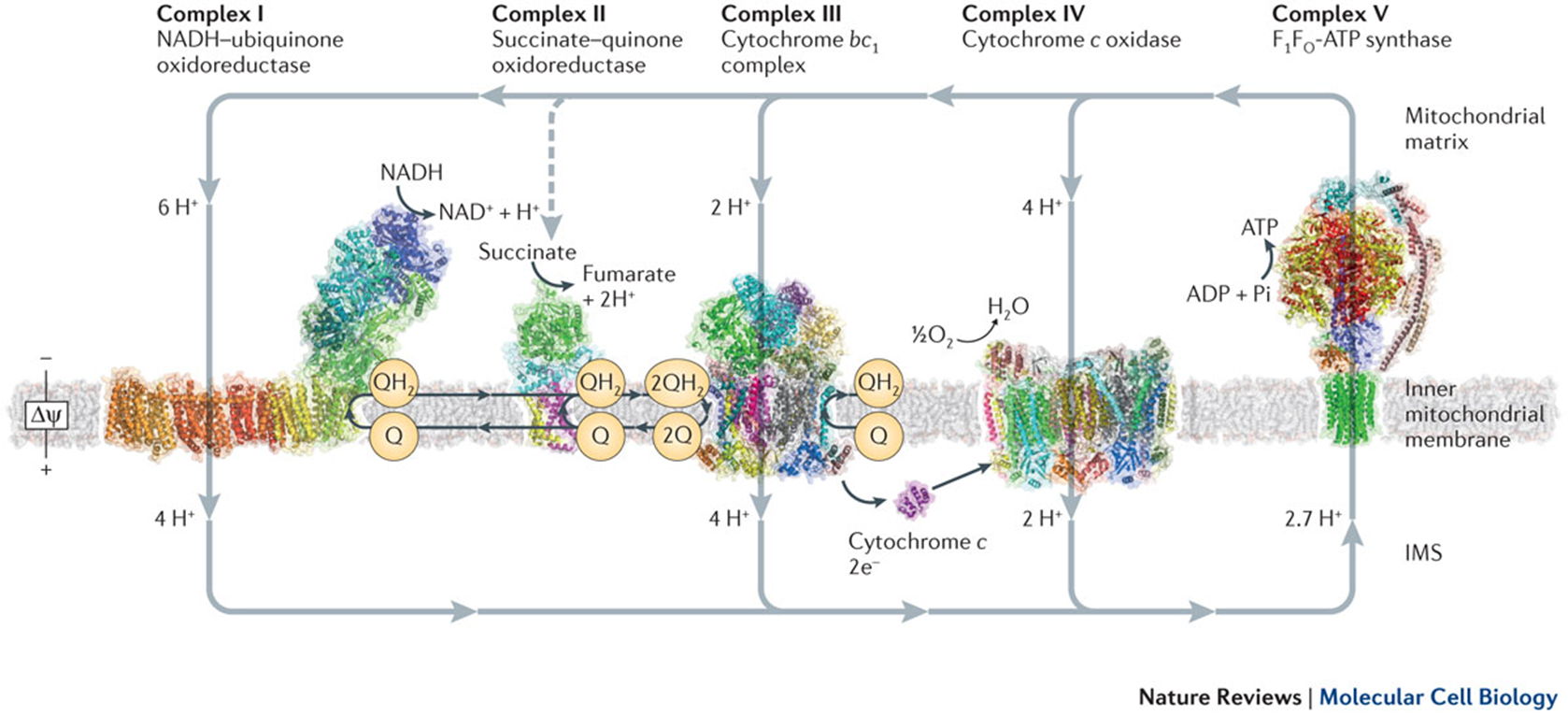
ATP generation: F1/F0 ATPase
At this point all the protons have been pumped across the membrane ready for ATP synthesis
F1 is the site of ATP synthesis
F0 is the motor unit
couples delta p driven protein rotation with ATP synthesis
coupling between F1 and F0 is done by the central stalk
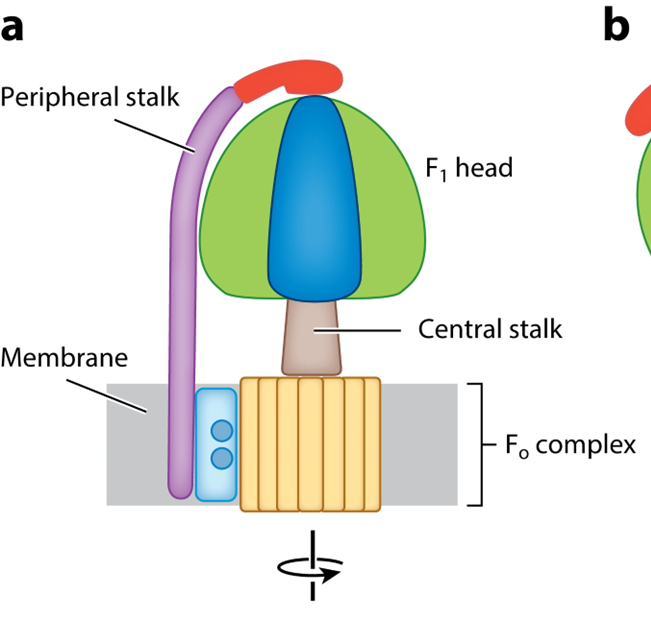
STRUCTURE
catalytic domain has an alpha and beta subunit
theres a stalk domain with a gamma delta and epsilon subunit
A C10 ring with an A subunit which is part of the motor complex
the peripheral stalk remains constant
catalytic domains oscillate which is coupled to ATP synthesis
F0 MOTOR
2 types of subunit:
A subunit: a stator which provides half channels for translocation of proteins across the bilayer
C subunit: a rotor, 8 copies in the mitochondria
couples the movement of protons across the bilayer to the rotation of the central stalk
the C subunits form a ring with a conserved glutamate in the middle
there are two half channels and the protos get transferred to the glutamate between them
the C subunits form a ring of charge and as the rotor rotates the protons get bought round to the second half channel
they get removed from the Glu by arginine
the number of C subunits varies across organisms
the lower the electrochemical potential requires more protons to move across the bilayer
as a result it puts more C subunits into the bilayer which moves more low energy protons across to generate ATP
transmission to F1 via electrical stalk
processive - as it rotates it pushes against the things that are around it which provides energy for ATP generation
3 roughly symetrical domains
3 a/b domains
1 ATP
1 ADP + Pi
1 is empty
the gamma subunit distorts the binding site which pushes the ADP + Pi
as the stalk rotates it causes a conformational change in the subunits which causes it to be converted from ADP —> ATP
whilst the stalk rotates the bound ATP undergoes a conformational shape change which leads to the release of ATP
the same conformational shape change causes the binding site to collapse down round the ADP and Pi
this allows ADP and Pi to bind to the binding site so it gets converted to ATP
each rotation generates 3ATP