BIOL 3301 Ch 14: Gene Regulation in Bacteria
1/72
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
73 Terms
gene regulation
level of gene expression can vary under different conditions
constitutive genes
unregulated genes
have essentially constant levels of expression
frequently code for proteins that are continuously necessary for the survival of the organism
(it’s fine that they’re unregulated bc the benefit of regulating genes is that coded proteins will be produced only when required)
only when required
the benefit of regulating genes is that coded proteins will be produced ?
constitutive genes
gene expression always “on”
unregulated
regulated genes
gene expression sometimes “on”
ch 14 bacteria vs ch 15 eukarotes
how do proteins that bind to DNA regulatory sequences somehow manage to either increase or decrease the rate of transcription by RNA polymerase?
?
Transcription factors control gene expression.
Repressors block transcription (off).
Activators help transcription (on).
Example: Lac operon regulates gene.
?
DNA is inside the nucleus.
Enhancers increase transcription (help).
Silencers decrease transcription (block).
Additional proteins fine-tune regulation.
gene regulation
? is important for these cellular processes
metabolism
response to environmental stress
cell division
regulation can occur at ANY of the points on the pathway to gene expression
there are conditions under which some genes are expressed and other conditions under which they are not
common gene expression regulation points in bacteria
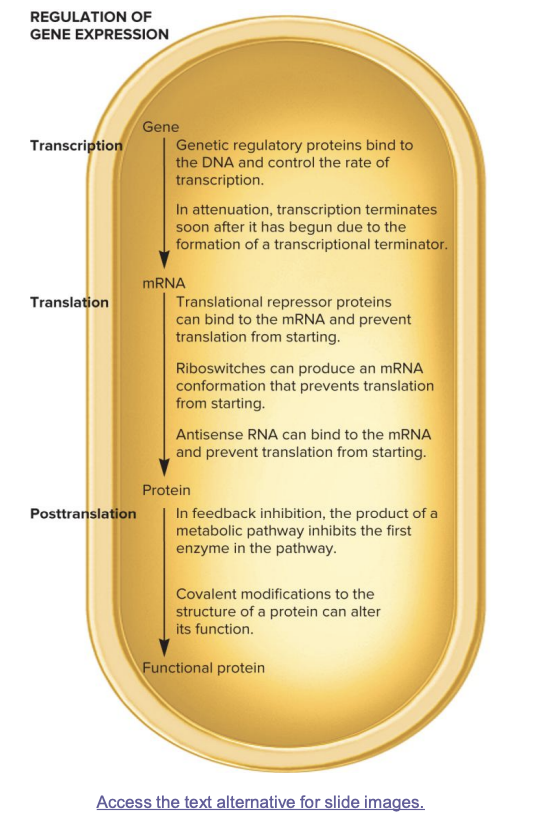
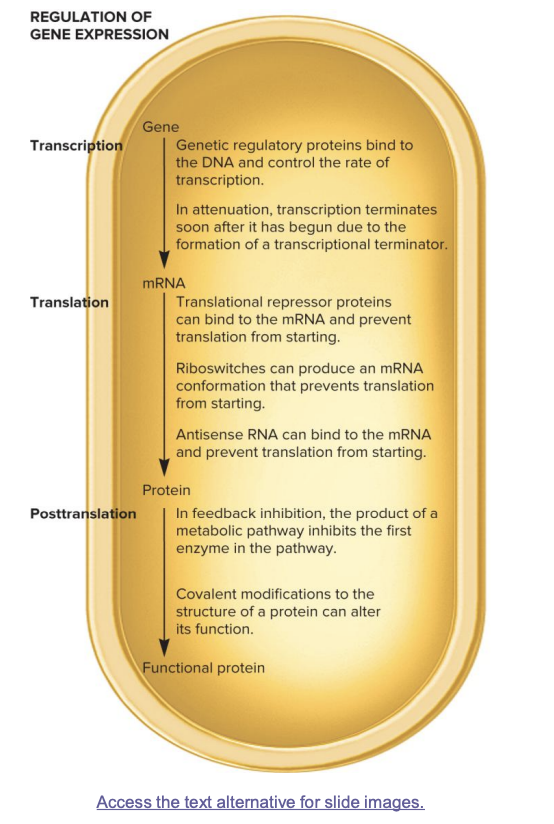
transcription (gene→mRNA)
genetic regulatory proteins bind DNA and control transcription rate
in attenuation, transcription is terminated soon after it starts due to formation of transcriptional terminator
translation (mRNA→ protein)
translational repressor proteins bind mRNA and prevent translation from starting
riboswitches produce an mRNA conformation preventing translation from starting
antisense RNA binds mRNA and prevents translation from starting
posttranslation (protein→ functional protein)
in feedback inhibition, the product of a metabolic pathway inhibits the first enzyme in the pathway
covalent modification to the structure of a protein can alter its function
transcription gene regulation in bacteria
genetic regulatory proteins bind to the DNA and control the transcription rate
Lac Operon: The lac repressor blocks transcription when lactose is absent. When lactose is present, it binds to the repressor, allowing RNA polymerase to transcribe the genes needed for lactose metabolism.
Trp Operon: The trp repressor binds to the operator region and prevents transcription when tryptophan is abundant. When tryptophan is scarce, the repressor detaches, allowing transcription for tryptophan biosynthesis.
occurs when gene is being copied into mRNA
posstranslational gene regulation in bacteria
in feedback inhibition, the product (final) of a metabolic pathway inhibits the first enzyme pathway
prevents the pathway from producing more of the product than needed
covalent modification to the structure of a protein can alter its function
e.g., phosphorylation, acetylation, methylation, ubiquitination,
modifications change protein’s shape, activity, stability or localization
ex., phosphorylation can activate/deactivate enzymes by altering their conformation
ubiquitination may mark some proteins for degradation
regulation can occur as protein is undergoing changes to become a functional protein
transcription initiation
the most common way to regulate gene expression in bacteria
the rate of RNA synthesis can be increased or decreased
involves regulatory transcription factors (RTFs)← proteins!
repressors: bind DNA and inhibit transcription
activators: bind DNA and increase transcription
regulatory transcription factors RTFs
proteins that regulate transcription
repressors: bind DNA and inhibit transcription
activators: bind DNA and increase transcription
repressors
regulatory transcription factors RTFs that bind DNA and inhibit transcription
activators
regulatory transcription factors RTFs that bind DNA and increase transcription
negative control
transcriptional regulation by repressor proteins
gene expression on→ (repressor protein action)→ off
positive control
transcriptional regulation by activator proteins
gene expression: off → (activator protein action)→ on
small effector molecules
? affect transcription regulation
bind to regulatory transcription factors but not to DNA directly
may increase or decrease transcription
inducers
increase transcription
bind activators and cause them to bind DNA
bind to repressors and prevent them from binding to DNA
genes regulated in this manner are inducible
may inhibit transcription
corepressors bind repressors and cause them to bind DNA
inhibitors bind activators and prevent them from binding DNA
genes regulated in this manner are repressible
inducers
small effector molecule that increases transcription
bind to regulatory transcription factors but not DNA directly
bind activators and cause them to bind DNA
bind repressors and prevent them from binding to DNA
genes regulated in this manner are inducible
inducible genes
regulated by small effector molecules (inducers)
inducers increase transcription
bind to regulatory transcription factors but not DNA directly
bind activators and cause them to bind DNA
bind repressors and prevent them from binding to DNA
corepressors
bind repressors and cause them to bind to DNA
small effector molecule that inhibits transcription
regulate repressible genes
inhibitors
bind activators and prevent them from binding DNA
small effector molecule that inhibits transcription
regulate repressible genes
repressible genes
regulated by small effector molecules that inhibit transcription
corepressors bind repressors and cause them to bind DNA
inhibitors bind activators and prevent them from binding DNA
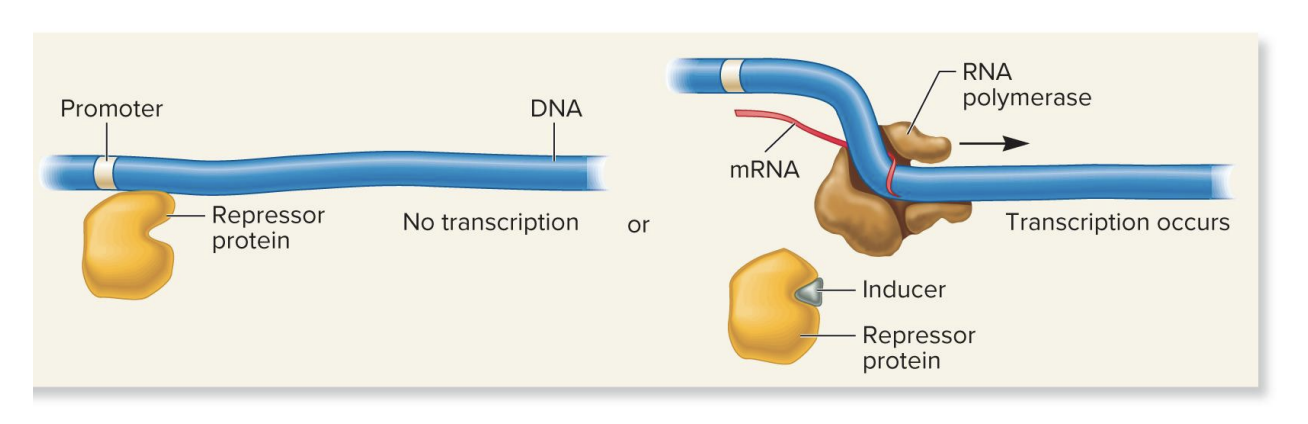
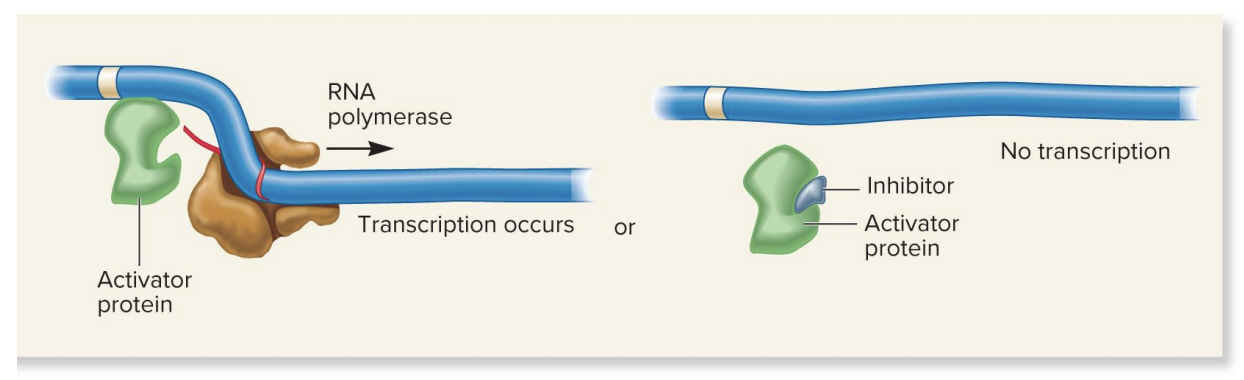
repressor protein, inducer molecule, inducible gene
no inducer present
repressor protein blocks transcription
inducer present
inducer binds repressive to repressor protein, causing a conformation change that prevents the repressor from binding to the DNA, thereby allowing transcription to proceed
activator protein, inducer, inducible gene
no inducer present
activator protein can’t bind DNA, transcription does not occur
inducer present
inducer binds activator protein, allowing it to bind to the DNA and activate transcription
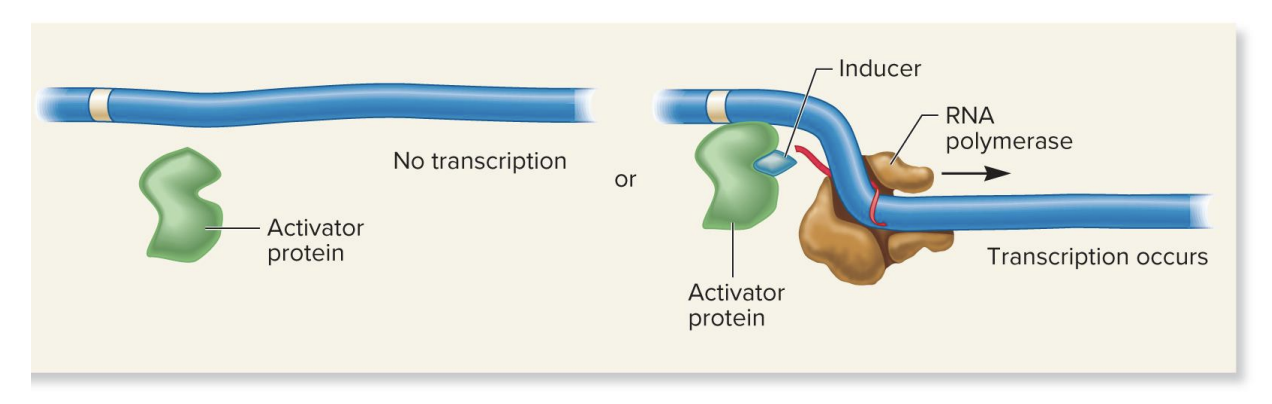
repressor protein, corepressor molecule, repressible gene
no corepressor present
repressor protein won’t bind DNA→ transcription occurs
corepressor present
corepressor binds repressor protein, causing a conformation change that allows the repressor to bind the DNA→ inhibit transcription
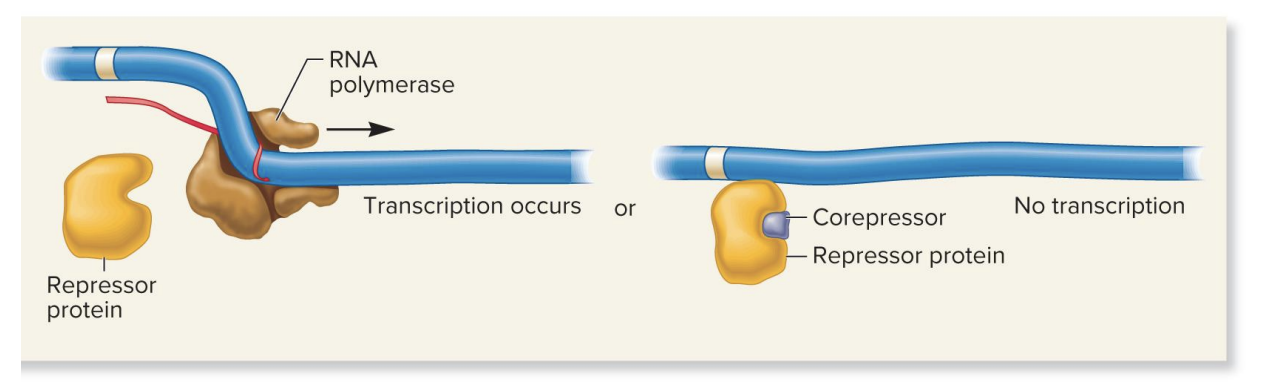
activator protein, inhibitor molecule, repressible gene
no inhibitor present
activator protein binds DNA without needing an effector molecule→ transcription proceeds
inhibitor present
inhibitor binds to activator protein, causing a conformational change that prevents the activator from binding to the DNA→ transcription inhibited
enzyme adaptation
a particular enzyme appears within a living cell only after the cell has been exposed to the substrate for that enzyme
lactose metabolism
in 20th century, Francois Jacob and Jacques Monod at Pasteur Institute in Paris were interested in the phenomenon of enzyme adaptation
particular enzyme appears in cell only after cell has been exposed to enzyme’s substrate
their focus was on ? in E. coli
observed
medium w/o lactose
no detectable presence of beta-galactosidase in E. coli cell
medium w/ lactose
lots of beta-galactosidase in E. coli cell
why?
seemed like the cell could "detect" the presence of lactose (its substrate) and began to produce the enzyme (beta galactosidase) in response.
However, lactose isn't the cell's "favorite food," so the cell produces the enzyme only when lactose is present, indicating inducible gene regulation
wo lactose
E coli grown in ? medium
does not produce the enzyme β-galactosidase (an enzyme needed to metabolize lactose)
seemed like the cell could "detect" the presence of lactose (its substrate) and began to produce the enzyme (beta galactosidase) in response.
However, lactose isn't the cell's "favorite food," so the cell produces the enzyme only when lactose is present, indicating inducible gene regulation
w lactose
E coli grown in ? medium
cell starts producing large amounts of β-galactosidase.
a
operon
regulatory unit consisting of a few protein-coding genes under control of 1 promoter
codes a polycistronic mRNA
contains coding sequence for 2 or more protein-coding genes
allows bacterium to coordinately regulate a group of genes that code proteins with a common functional goal
E. coli genes involved in lactose utilization have 2 transcriptional uniits
Lac Operon: One transcriptional unit that includes the genes responsible for lactose metabolism (e.g., lacZ, lacY, lacA).
lacI Gene: A separate transcriptional unit that codes for the lac repressor protein, which controls the lac operon.
2 transcriptional units
genes in E coli involved in lactose utilization have ?
lac operon
One transcriptional unit that includes the genes responsible for lactose metabolism (e.g., lacZ, lacY, lacA)
regulatory unit that codes for proteins involved in lactose metabolism
DNA elements that control transcription
promoter→ bind RNA polymerase
operator→ bind lac repressor protein
CAP site→ binds the catabolite activator protein CAP
terminator→ ends transcription
protein coding genes
lacZ
codes beta galctosidase
enzymatically cleaves lactose and lactose analogs
also converts lactose→ allolactose (an isomer)
lacY
codes lactose permease
membrane protein required for transport of lactose and analogues
lacA
codes galactosidase transacetylase
covalently modifies lactose and analogues
prevents toxic buildup of nonmetabolizable lactose analogs
lacl gene
A separate transcriptional unit that codes for the lac repressor protein, which controls the lac operon.
not considered part of lac operon
has its own promoter, i promoter
constitutively expressed at fairly low intervals
codes lac repressor
lac repressor protein fxs as a tetramer
only a small amount of protein is needed to repress the lac operon
lac operon
One transcriptional unit that includes the genes responsible for lactose metabolism (e.g., lacZ, lacY, lacA) in E coli
regulatory unit that codes for proteins involved in lactose metabolism
(its actual parts are) DNA elements that control transcription
promoter→ bind RNA polymerase
operator→ bind lac repressor protein
CAP site→ binds the catabolite activator protein CAP
terminator→ ends transcription
protein coding genes
lacZ
codes beta galctosidase
enzymatically cleaves lactose and lactose analogs
also converts lactose→ allolactose (an isomer)
lacY
codes lactose permease
membrane protein required for transport of lactose and analogues
lacA
codes galactosidase transacetylase
covalently modifies lactose and analogues
prevents toxic buildup of nonmetabolizable lactose analogs
lac promoter
bind RNA polymerase
part of lac operon
operator site laco
sequences of bases that bind lac repressor protein, which blocks transcription when lactose is absent.
part of lac operon
CAP site
DNA sequence recognized by/binds an activator protein the catabolite activator protein CAP
part of lac operon
lac terminator
ends transcription
part of lac operon
lac z
codes beta galactosidase
enzymatically cleaves lactose and lactose analogs
also converts lactose→ allolactose (an isomer)
part of lac ooperon
lacY
codes lactose permease
membrane protein required for transport of lactose and analogues
part of lac operon
lacA
codes galactosidase transacetylase
covalently modifies lactose and analogues
prevents toxic buildup of nonmetabolizable lactose analogs
part of lac operon
lacl gene
A separate transcriptional unit that codes for the lac repressor protein, which controls the lac operon.
not considered part of lac operon
has its own promoter, i promoter
constitutively expressed at fairly low intervals
codes lac repressor
lac repressor protein fxs as a tetramer
only a small amount of protein is needed to repress the lac operon
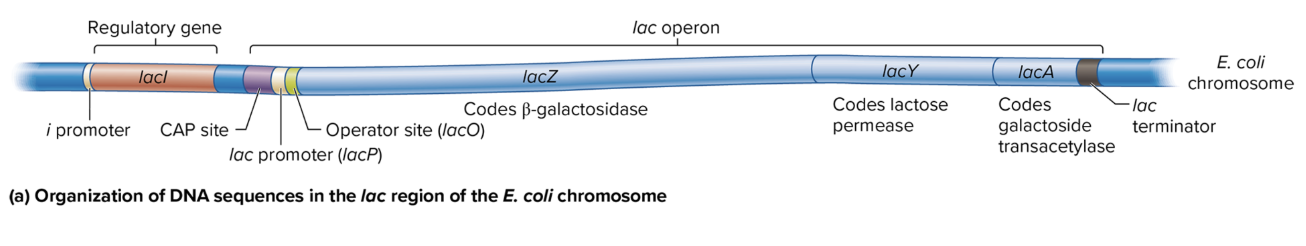
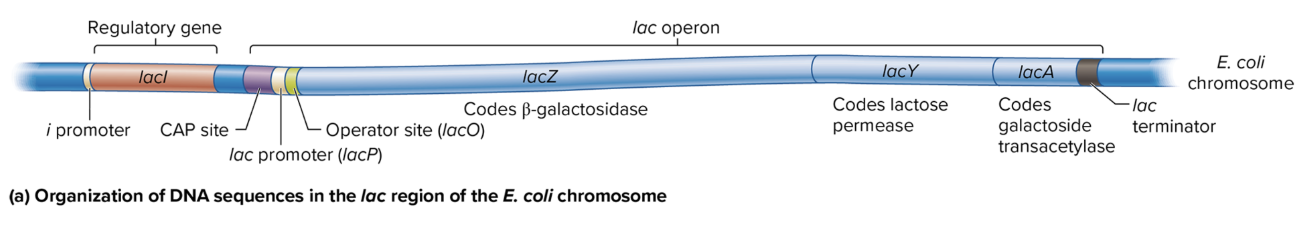
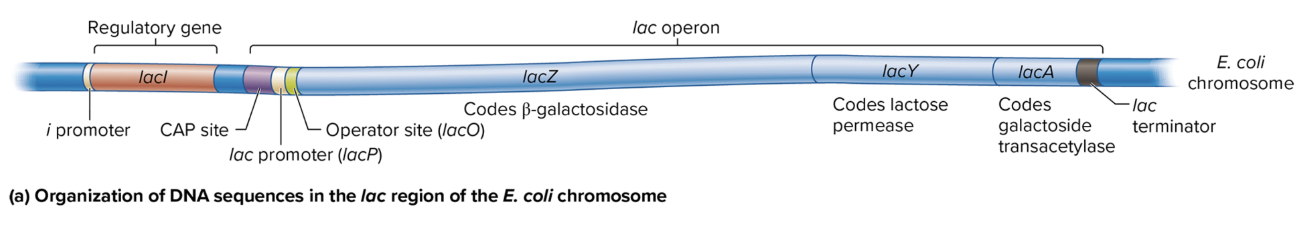
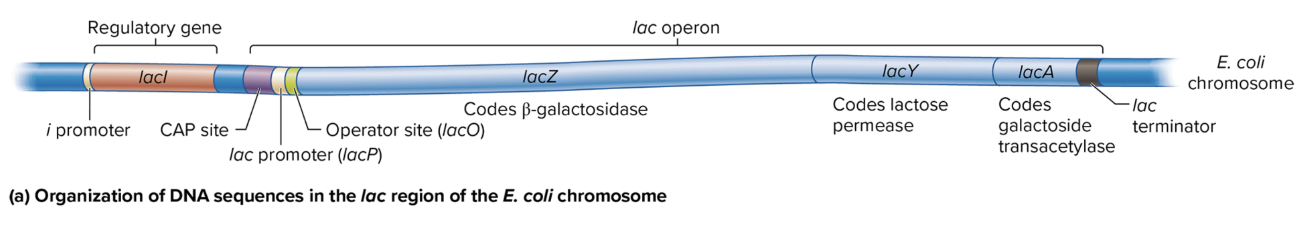
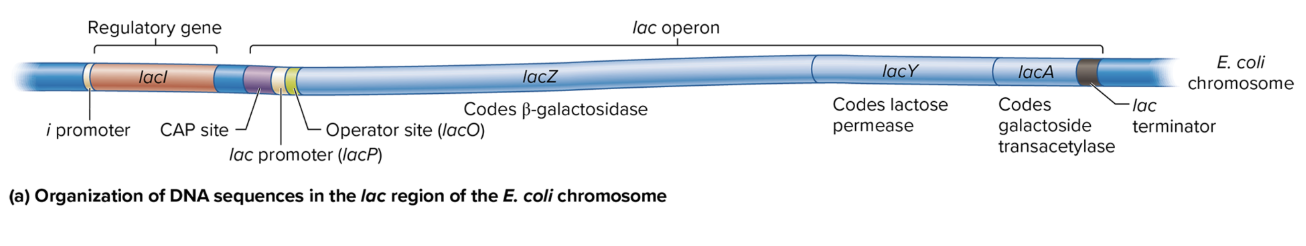
DNA sequence organization in lac region of E coli chromosome
i Promoter: Drives transcription of the lacI gene (codes for the lac repressor).
CAP Site: Binds the catabolite activator protein (CAP) to help activate the operon when glucose is low.
lac Promoter (lacP): Controls the transcription of the lacZ, lacY, and lacA genes as a single polycistronic mRNA.
Operator Site (lacO): Binds the lac repressor protein, which blocks transcription when lactose is absent.
lac Terminator: Marks the end of transcription for the lac operon.
lacZ: Codes for β-galactosidase, which breaks down lactose into glucose and galactose.
lacY: Codes for lactose permease, which allows lactose entry into the cell.
lacA: Codes for galactoside transacetylase, involved in detoxifying certain sugars.
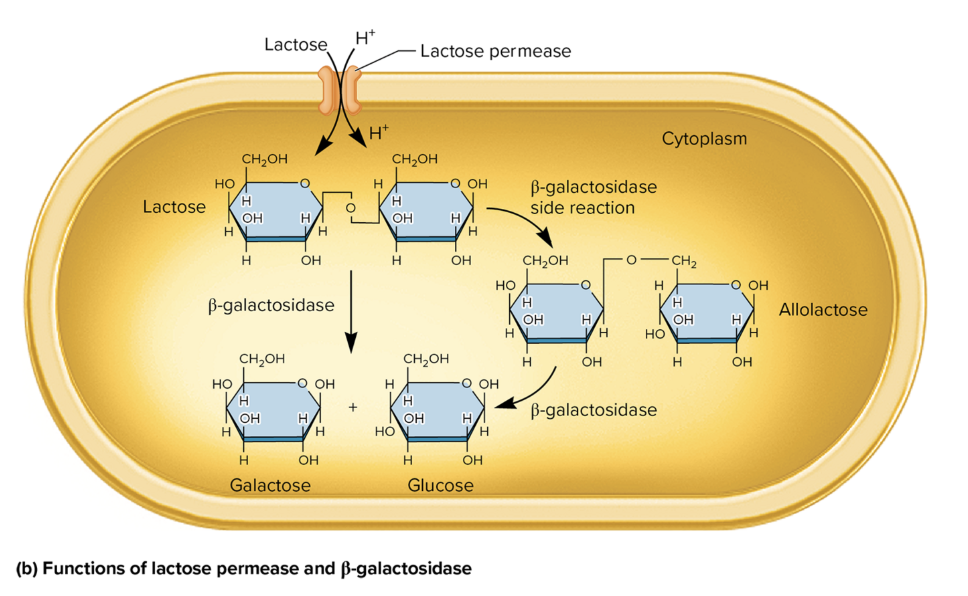
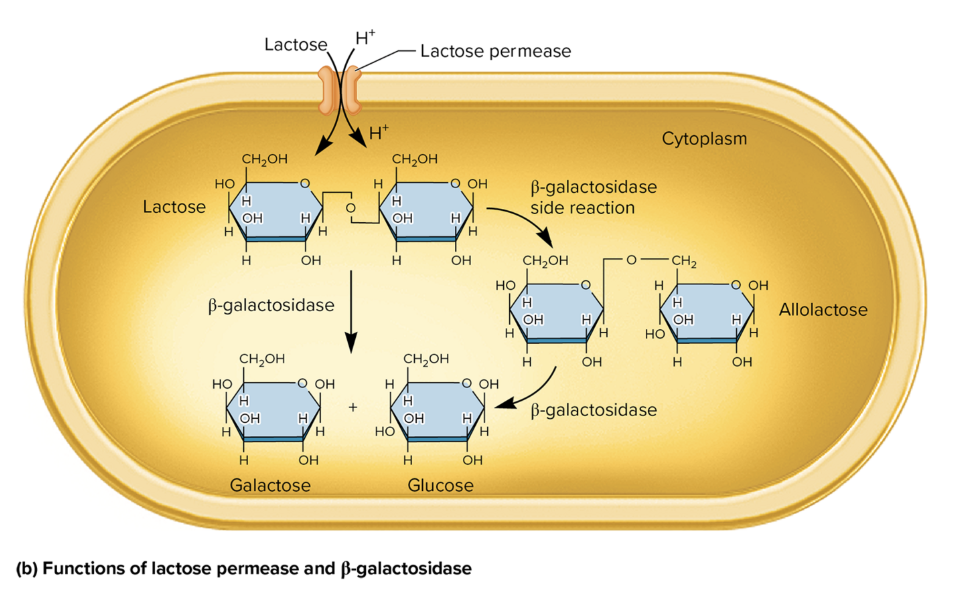
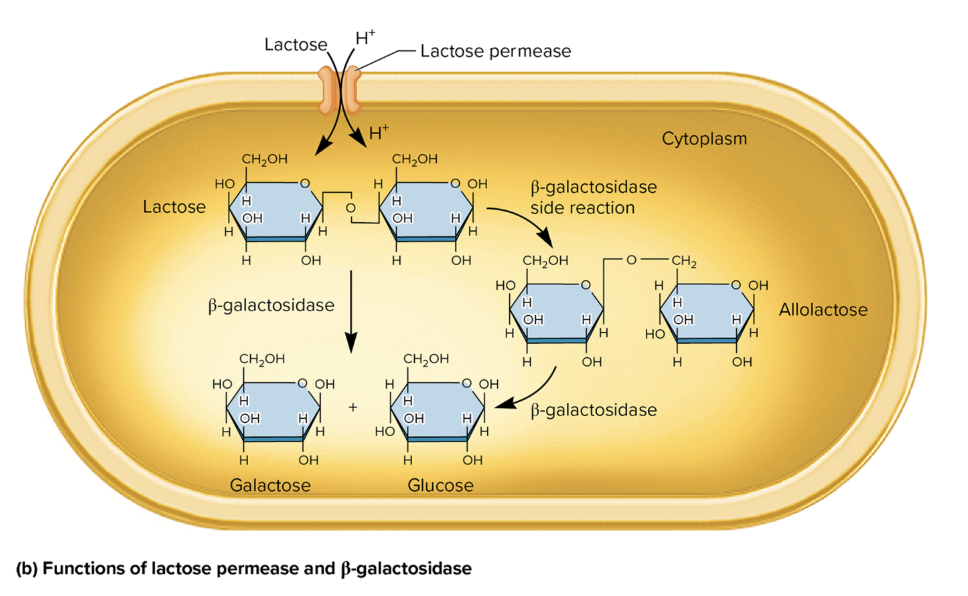
protein fx in lactose metabolism
lactose permease
facilitates active transport of lactose→ cytoplasm
use proton gradient to co-transport lactose and hydrogen ions (H+)
β-Galactosidase
break down lactose→ glucose + galactose
catalyzes a side rxn, converting lactose→ allolactose (an inducer of the lac operon)
allolactose is further broken down→ glucose and galactose via β-galactosidase.
lactose permease
protein
facilitates active transport of lactose→ cytoplasm
use proton gradient to co-transport lactose and hydrogen ions (H+)
beta galactosidase
break down lactose→ glucose + galactose
catalyzes a side rxn, converting lactose→ allolactose (an inducer of the lac operon)
allolactose is further broken down→ glucose and galactose via β-galactosidase.
protein
lac operon
lac operon
? can be transcriptionally regulated
by repressor protein (negative control)
by activator protein (positive control)
Inducible, negative control mechanism
lac repressor protein
inducer is allolactose (regulation)
acts like an inducer that inactivates the lac repressor via allosteric regulation
In absence of lactose in environment
inducer allolactose is also absent
lac repressor/repressor protein is tightly bound to the operator site (lacO), preventing RNA polymerase from transcribing the lac operon
when lactose is present in environment
inducer allolactose is available
lactose → (β-galactosidase)→ allolactose
allolactose binds repressor, altering the conformation of the repressor protein, which prevents i from binding the operator site (lacO)
this allows RNA polymerase to transcribe the lac operon genes, enabling lactose metabolism
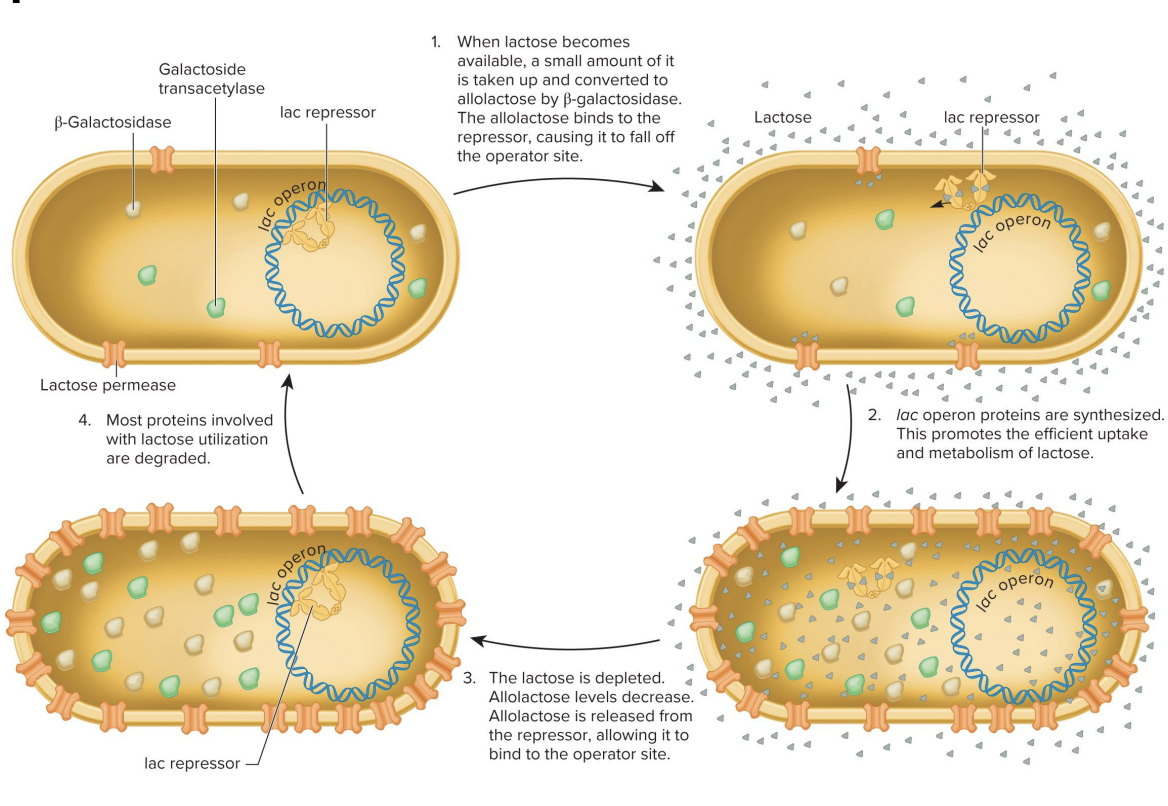
cycle of induction and repression
ensures lac operon is only active when lactose is available, allowing efficient regulation of lactose metabolism in E.coli
lactose absent
inducer allolactose is also absent
lac repressor/repressor protein is tightly bound to the operator site (lacO), preventing RNA polymerase from transcribing the lac operon
lactose present
inducer allolactose is available
lactose → (β-galactosidase)→ allolactose
allolactose binds repressor, altering the conformation of the repressor protein, which prevents it from binding the operator site (lacO)
this allows RNA polymerase to transcribe the lac operon genes, enabling lactose metabolism
induction and repression cycle
lactose available
small amount lactose→ (via β-galactosidase) → allolactose
allolactose binds repressor, causing it to fall of the operator site (change conformation) which prevents it from binding the operator site (lacO)
RNA polymerase transcribes lac operation genes
lac operon proteins are synthesized
promotes efficient uptake and lactose metabolism
lactose is depleted (lactose becomes more unavailable)
allolactose levels decrease
allolactose released from repressor, allowing it to bind to the operator site
more proteins involved with lactose utilization are degraded
lactose permease
beta galactosidase
lac repressor
galactoside transacetylase
purpose
ensures lac operon is only active when lactose is available, allowing efficient regulation of lactose metabolism in E.coli
lacl gene
In 1950s, Jacob, Monod, and Pardee ID’ed mutant strains in bacteria with abnormal lactose adaptation
lacl gene mutation: (lacl-)
defect in the lacl gene
led to constitutive expression of lac operon, even when lactose was absent
basically the continuous expression of the lac operon→ always producing the enzymes needed for lactose metabolism
these mutations were mapped very close to the lac operon on the bacterial chromosome
bacterial conjugation
researchers used bacterial conjugation to introduce different portions of the lac operon onto different strains
F’ factors (plasmids) were used to transfer parts of the lac operon, such as the lacl gene, into recipient bacteria
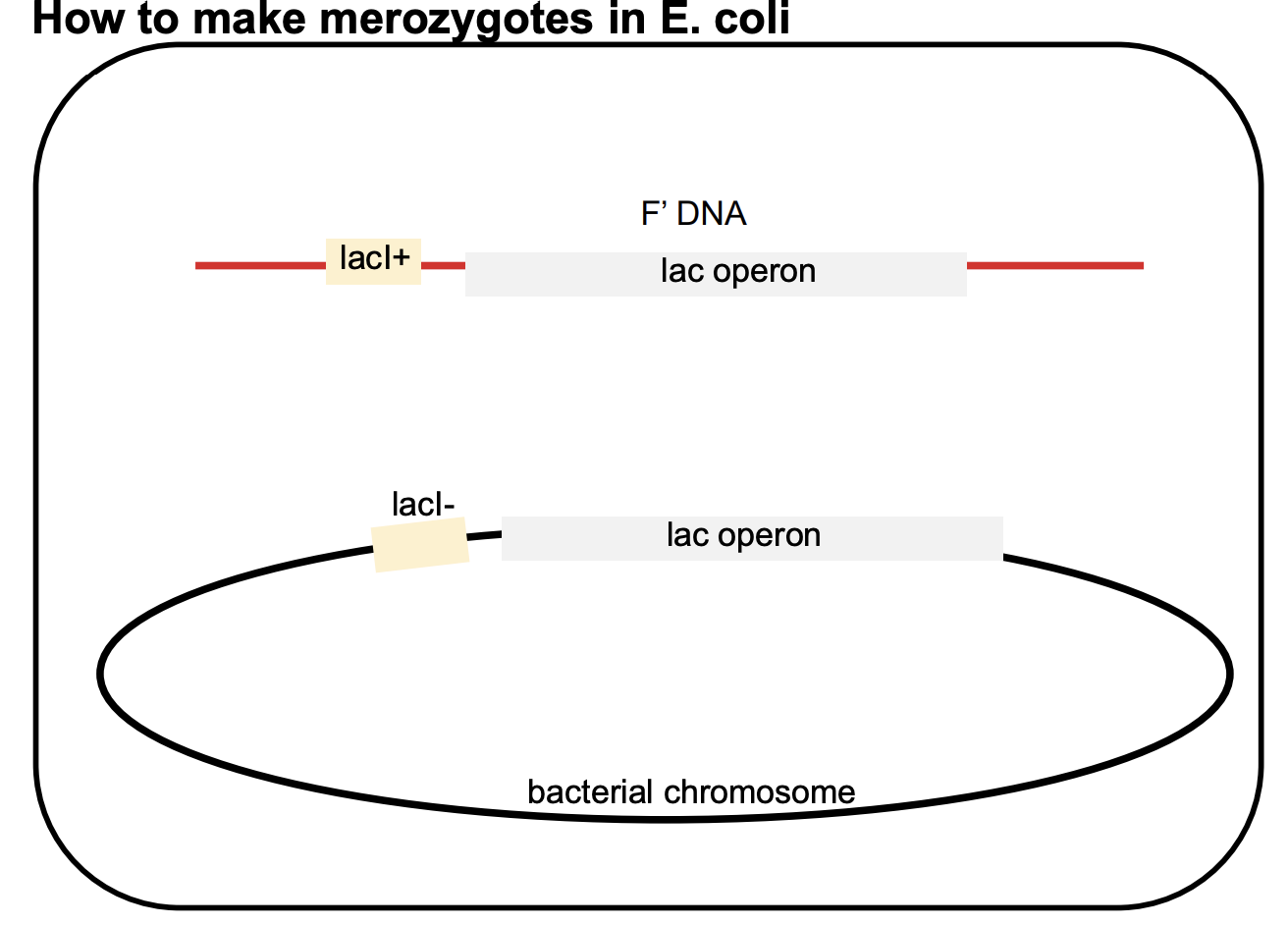
merozygotes (partial diploids)
after transferring the lacl gene via F’ factors, bacteria could have 2 copies of the lacl gene: one on the chromosome and one on the plasmid (F’ factor)
these bacteria are called merozygotes/partial diploids bc they have 2 copies of a specific gene, resulting in a more complex genetic makeup
lacl-
defect in the lacl gene
led to constitutive expression of lac operon, even when lactose was absent
basically the continuous expression of the lac operon→ always producing the enzymes needed for lactose metabolism
these mutations mapped very close to the lac operon
bacterial conjugation
researchers used bacterial conjugation to introduce different portions of the lac operon onto different strains
F’ factors (plasmids) were used to transfer parts of the lac operon, such as the lacl gene, into recipient bacteria
merozygotes/partial diploids
? instrumental in allowing Jacob, Monod, and Pardee to elucidate the fx of the lacI gene
2 key points
the 2 lacI genes in a ? may be different alleleles
lacI− (mutant allele) on the bacterial chromosome.
lacI+ (wild-type allele) on the F’ factor (plasmid).
genes on the F’ factor (plasmid) are not physically connected to those on the bacterial chromosome
allows the study of how two copies of the lacI gene can interact.
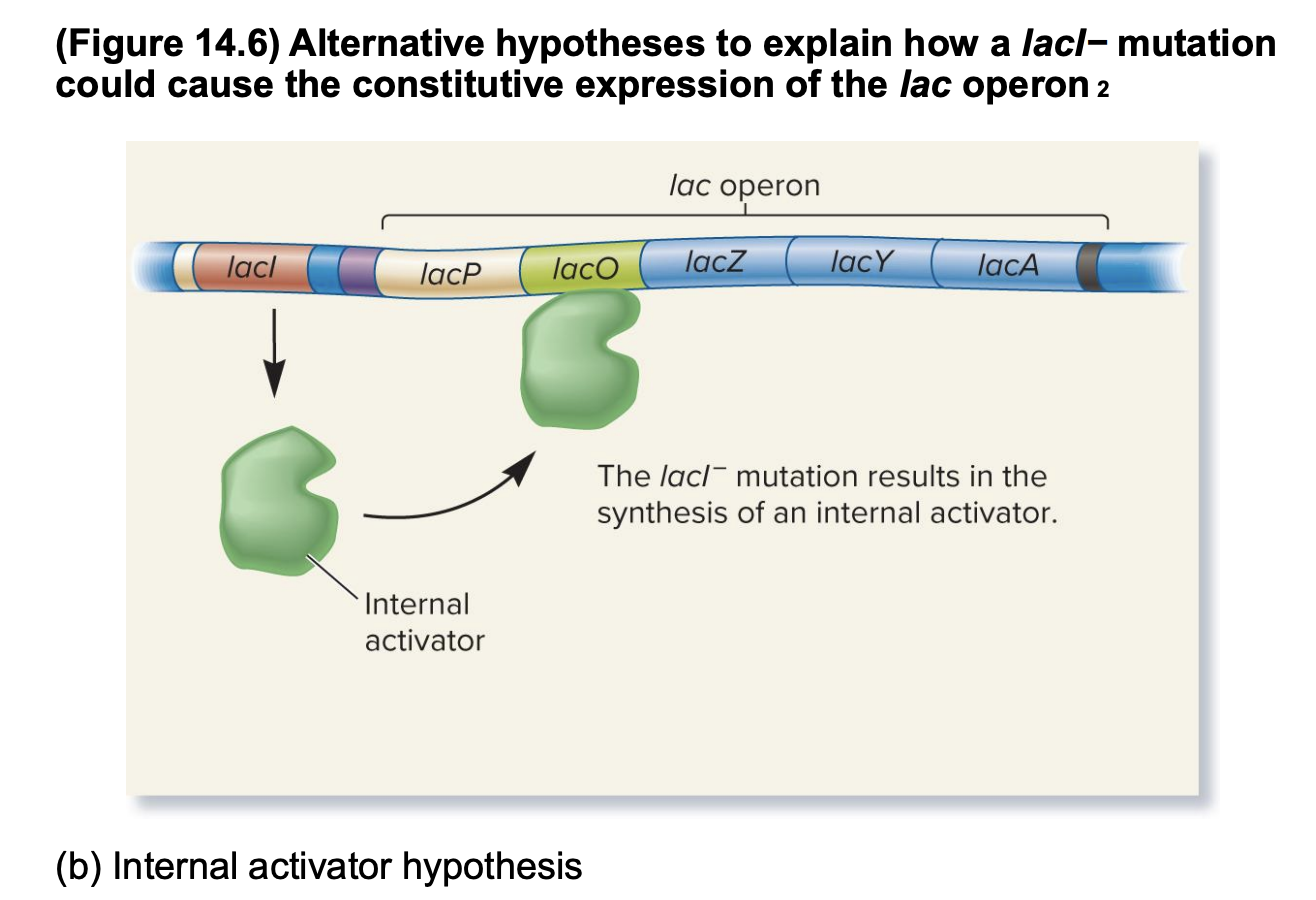
internal activator hypothesis
Jacob, Monod, and Pardee hypothesized that the lacI- mutation results in the synthesis of an internal inducer
internal activator binds to the lacO operator, which activates the transcription of the lac operon, causing constitutive expression
e continuous or unregulated expression of a gene or operon
if correct: the inducer protein produced from the chromosome can diffuse and activate the lac operon on the F’ factor
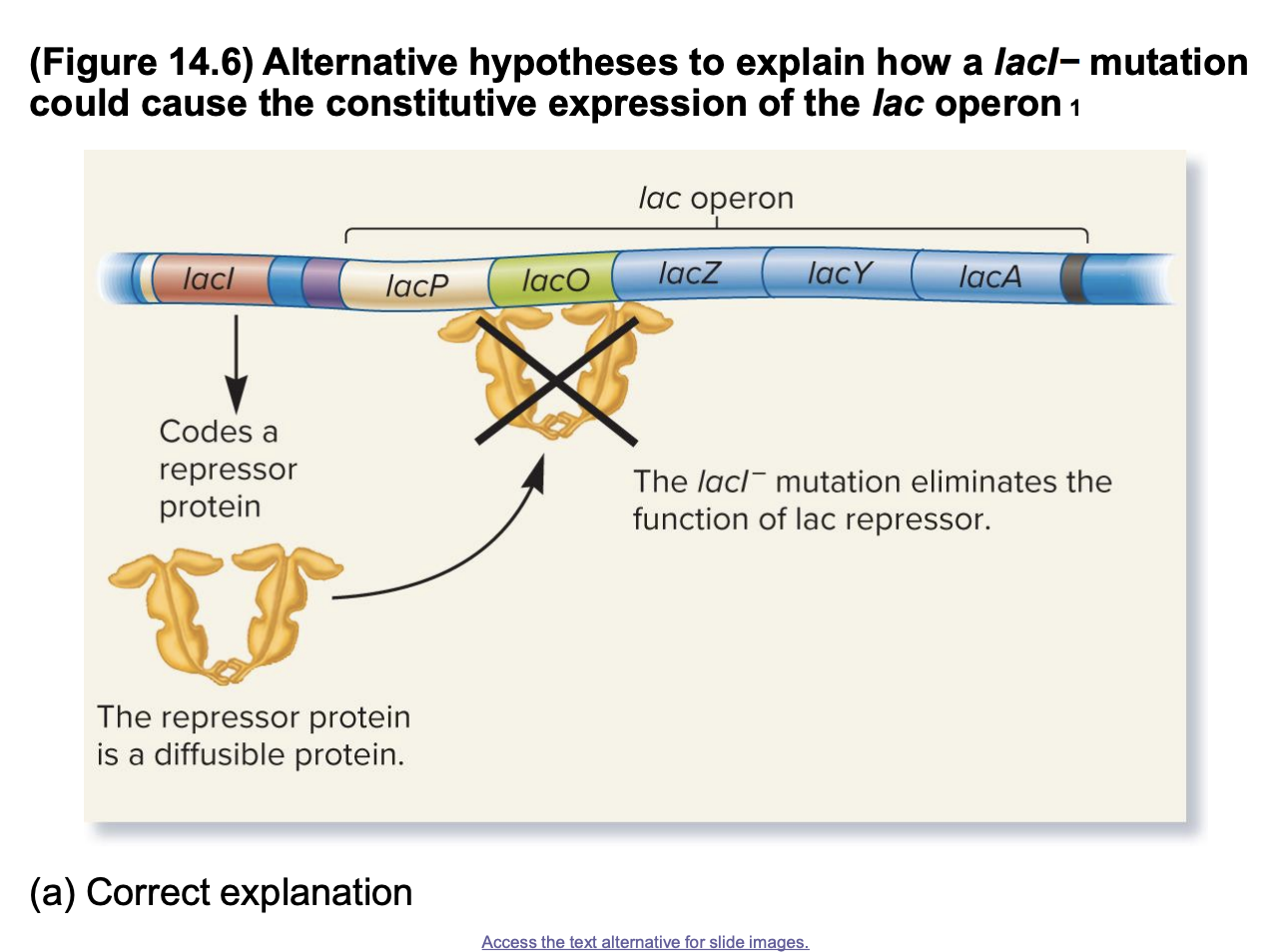
alternative hypothesis
the lacI- mutation eliminates the fx of a lac repressor that can diffuse throughout the cell
The lac repressor is unable to bind to the lacO operator, allowing RNA polymerase to transcribe the lac operon continuously, regardless of lactose presence
if correct: the repressor coded on the F’ factor can diffuse and turn off the lac operon on the bacterial chromosome
ability of the repressor to diffuse is important bc even when there is a mutated lacI gene on the chromosome (lacI−), the repressor on the F' factor (plasmid) can still function properly to regulate the lac operon and prevent its constitutive expression.
the correct expression
hypothesis testing
grow mutant strain and merozygote (partial diploid) strain separately
Mutant strain: lacI- (no repressor gene) → Constitutive expression of the lac operon.
Merozygote strain: Has two copies of lacI gene, one on the bacterial chromosome (lacI-) and one on the F' plasmid (lacI+), introduced by conjugation.
divide each strain into 2 tubes
add lactose to one of the 2 tubes
incubate the cells long enough to allow lac operon induction
lyse the cells with a sonicator. This allows beta galactosidase to escape from the cells.
add β-o-nitrophenylgalactoside (β ONPG)
a colorless compound
if β-galactosidase is present→ cleave compound to produce galactose and o-nitrophenol (O-NP)→ yellow color
O- nitrophenol is yellow, the deeper the yellow color, the more β-galactosidase was produced
β-ONPG is a colorless compound that is cleaved by β-galactosidase into galactose and o-nitrophenol (O-NP).
If β-galactosidase is present, O-NP will be produced, which is yellow. The intensity of the yellow color will indicate the amount of β-galactosidase activity
incubate the sonicated cells to allow f β-galactosidase time to cleave β-o-nitrophenylgalactoside
measure the yellow color produced with a spectrophotometer
In the mutant strain (with lacI- mutation), β-galactosidase will be expressed constitutively, meaning yellow color will be produced even without lactose.
In the merozygote strain, the expression depends on whether the lacI+ or lacI- is present, with the presence of lactose allowing activation of the lac operon in the lacI+ strain.
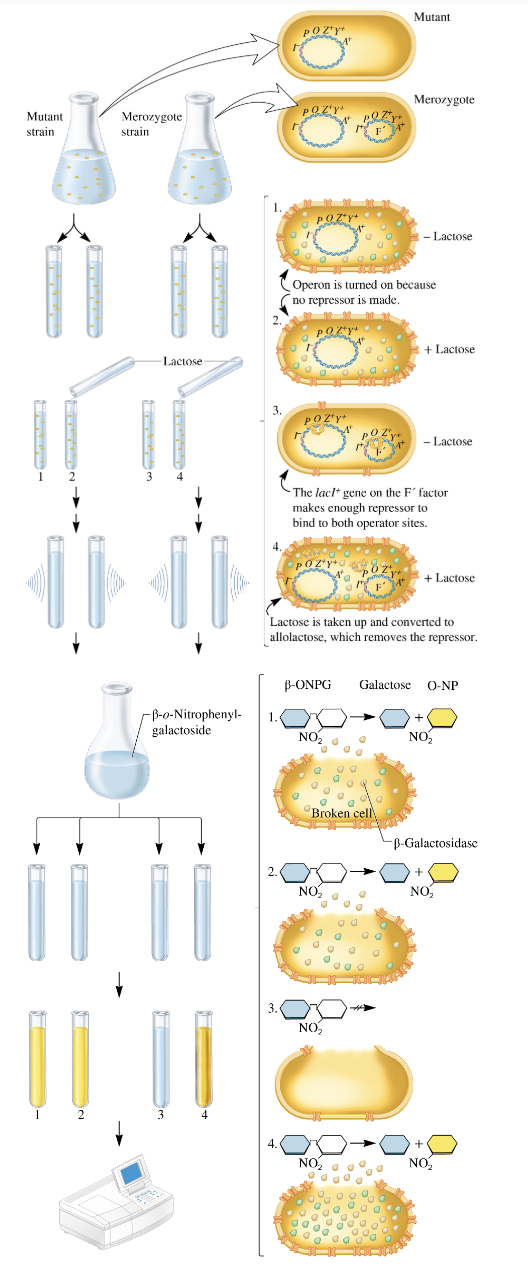
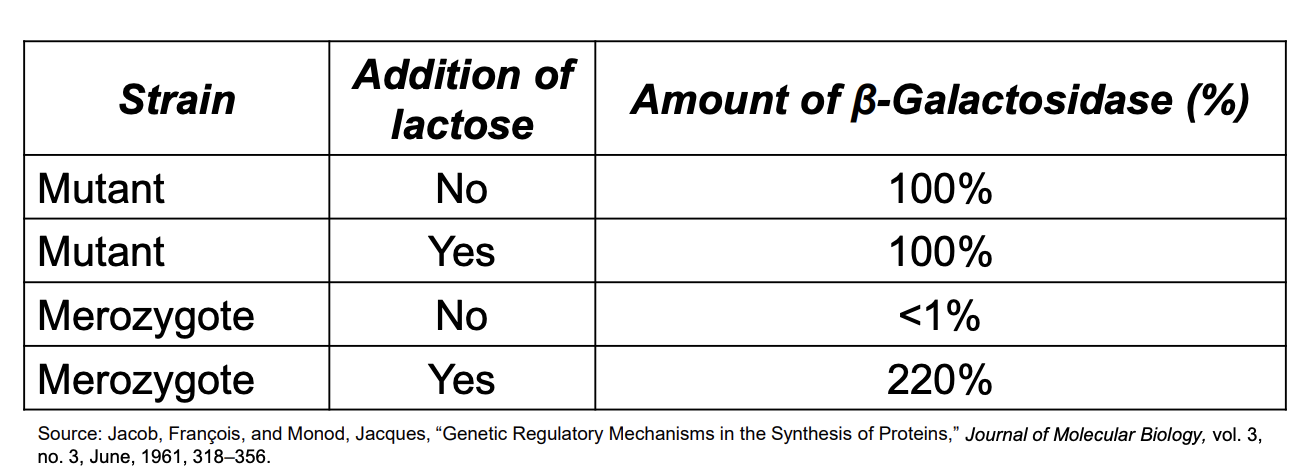
the data
mutant strains express the lac operon at 100% bc of the constitutive expression in the lacl- strain
merozygote
in the absence of lactose, both lac operons are repressed → <1%
in the presence of lactose, both lac operons are induced, yielding a higher level of enzyme activity→ 220%
therefore:
lacI- mutant doesn’t make a repressor protein
no repressor to bind to the operator (lacO) and block transcription→ continuous production of β-galactosidase, regardless of lactose present/absent
lacI+ (WT) gene makes a repressor protein that can repress the lac operon in the same cell.. doesn’t have to be on the same piece of DNA as its gene
produce a functional repressor that can regulate the lac operon, even if it's located on a different piece of DNA (e.g., the F' factor/plasmid). The repressor can regulate both operons in a merozygote
trans effect
interaction btw regulatory proteins and DNA sequences
genetic regulation that can occur even though DNA segments aren’t physically adjacent
mediated by genes that code regulatory transcription factors
ex., action of lac repressor on lac operon
lac repressor can bind the operator (lacO) of the lac operon to regulate the lac operon even though it’s located on a different piece of DNA (chromosome or F’ plasmid)
cis affect/cis acting element
DNA sequence that must be adjacent to the gene(s) it regulates
mediated by sequences that bind regulatory transcription factors
ex., the lac operator
lac operator (lacO) is DNA sequence near the lac genes (lacZ, lacY, lacA)
lacO is a binding site for the lac repressor protein
when lac repressor binds lacO, transcription of the lac operon is blocked
in presence of lactose or allolactose, the repressor detaches from lacO, allowing the lac operon to be transcribed.
mutation in trans-acting factor vs cis acting element
mutation in trans acting factor is complemented by the introduction of a second gene with normal function
can fix by introducing a second copy of the gene that codes for the normal protein
ex., if there is a lacI- mutation (which means the lac repressor is not made), introducing a normal lacI+ gene elsewhere can fix the mutation
mutation in a cis-acting element is not affected by the introduction of another normal cis-acting element
cannot be fixed by introducing another copy of the DNA sequence elsewhere, because the regulation has to happen right next to the gene it controls
ex., mutation in the lacO (operator) cannot be rescued by introducing a second lacO somewhere else because the repressor has to bind directly to the lacO that is adjacent to the operon
mutation in trans acting factor
complemented by the introduction of a second gene with normal function
can fix by introducing a second copy of the gene that codes for the normal protein
ex., if there is a lacI- mutation (which means the lac repressor is not made), introducing a normal lacI+ gene elsewhere can fix the mutation
mutation in cis-acting element
not affected by the introduction of another normal cis-acting element
cannot be fixed by introducing another copy of the DNA sequence elsewhere, because the regulation has to happen right next to the gene it controls
ex., mutation in the lacO (operator) cannot be rescued by introducing a second lacO somewhere else because the repressor has to bind directly to the lacO that is adjacent to the operon
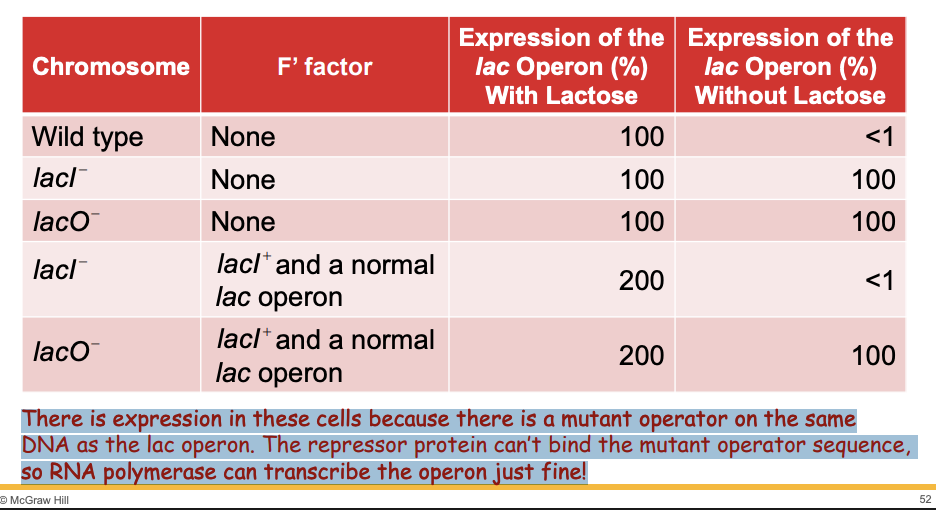
loss of fx mutation in lacI gene or operator site
Wild Type:
With Lactose: 100% expression (repressor inactivated by lactose).
Without Lactose: <1% expression (repressor binds operator).
lacI- (mutant):
With Lactose: 100% expression (no repressor).
Without Lactose: 100% expression (no repressor).
lacO- (mutant operator):
With Lactose: 100% expression (repressor can't bind).
Without Lactose: 100% expression (repressor can't bind).
lacI- and lacI+ on F' (merozygote):
With Lactose: 200% expression (activator protein boosts transcription).
Without Lactose: <1% expression (repressor binds operator).
lacO- and lacI- with normal lac operon:
With Lactose: 200% expression (activator protein boosts transcription).
Without Lactose: 100% expression (repressor can’t bind mutant operator).
Key Points:
lacI-: Constitutive expression (always on).
lacO-: No repressor binding, operon always transcribed.
Mutant operator (lacO-): The sequence has changed, so the repressor can't bind to it.
Result: Since the repressor can't block transcription, RNA polymerase is free to transcribe the lac operon, even in the absence of lactose.
Activator proteins (CAP) boost transcription when lactose is present.
catabolite repression
the lac operon can be transcriptionally regulated by an activator protein
glucose priority
when lactose and glucose are both available
E coli prefers to use glucose first
this prevents the lac operon from being activated and using lactose (? repression)
when glucose is depleted, ? repression is relieved, allowing the lac operon to be expressed (so bacteria can use lactose)
diauxic growth
the process where bacteria sequentially use 2 sugars (glucose first, then lactose)
diauxic growth
the process where bacteria sequentially use 2 sugars (glucose first, then lactose)
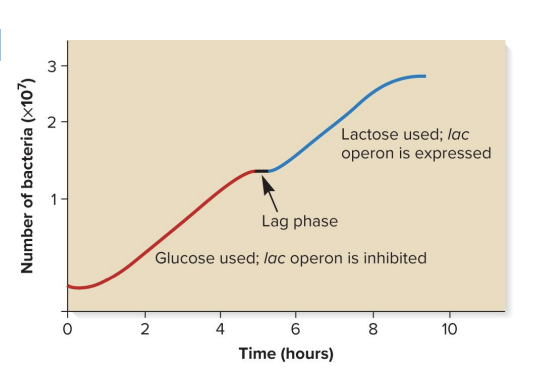
diauxic growth experiment
the process where bacteria sequentially use 2 sugars (glucose first, then lactose)
steps
E coli cells were given glucose and lactose at time zero
number of E. coli cells and concentrations of both extracellular glucose and lactose were monitored for 10 hours
findings
cells first used glucose to increase in number
glucose used: lac operon inhibited
so genes involved in lactose metabolism (lacZ, lacY, lacA) are not transcribed or expressed
catabolite repression keeps the lac operon off
after glucose was consumed a brief lag phase occurred as cells switched to utilization of lactose
lactose used: lac operon is expressed
so genes involved in lactose metabolism (lacZ, lacY, lacA) are transcribed and expressed
lag phase followed by a second increase in cell number until lactose was eventually depleted and growth leveled off
cyclic AMP cAMP
a small effector molecule involved in catabolite repression (hint: not glucose)
produced from ATP via enzyme adnenylyl cyclase
binds an activator protein known as the Catabolite Activator Protein CAP
tldr; low glucose→(E.coli uses ATP via adenylyl cyclase)→ higher cAMP levels
cAMP binds CAP→ cAMP-CAP complex→activate lac operon→ allowing transcription of lacZ, lacY, and lacA for lactose metabolization
remember; catabolite repression helping glucose preference/priority , (preventing the lac operon from being activated and using lactose) but now you’re low on glucose and need to switch to lactose
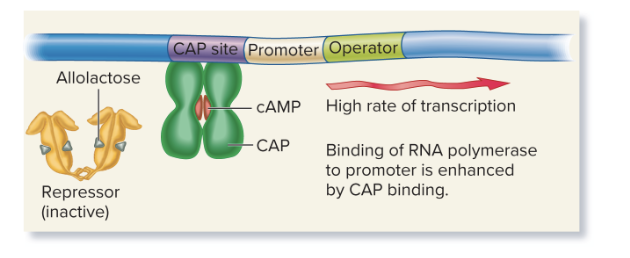
cAMP-CAP complex
an example of transcriptional regulation that is inducible and under positive control
binds CAP site near the lac promoter and increases transcription of lacZ, lacY, and lacA for lactose metabolization
in the presence of glucose, the enzyme adenylyl cyclase is inhibited
this decreases cAMP levels in the cell
(e.coli uses ATP via adenylyl cyclase to make cAMP)
cAMP no longer available to bind CAP
transcription rate of genes for lactose metabolism decreases
you now have glucose, no need use lactose (you prefer glucose)
lactose, no glucose (high cAMP)
High transcription: cAMP binds to CAP, enhancing RNA polymerase binding to the promoter, leading to high transcription (lacZ, lacY, and lacA for lactose metabolization).
Inactive repressor: Allolactose (inducer) binds to the repressor, preventing it from binding to the operator, allowing transcription.
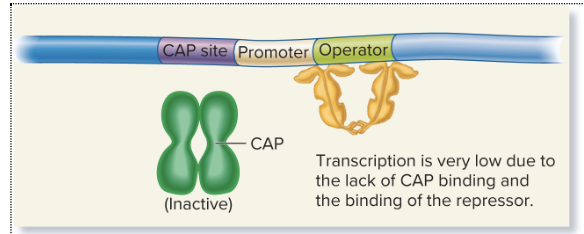
no lactose or glucose (high cAMP)
Low transcription: cAMP is still high, but the repressor remains active and blocks transcription (of lacZ, lacY, and lacA for lactose metabolization) at the operator, even though CAP is present.
Glucose levels are a key factor in determining cAMP levels. When glucose is absent, adenylyl cyclase (the enzyme that produces cAMP) is not inhibited, leading to high cAMP production
transcription is low because the lac repressor is still active (since lactose is absent, and thus there's no allolactose to inactivate the repressor). The repressor binds to the operator, blocking RNA polymerase and preventing transcription
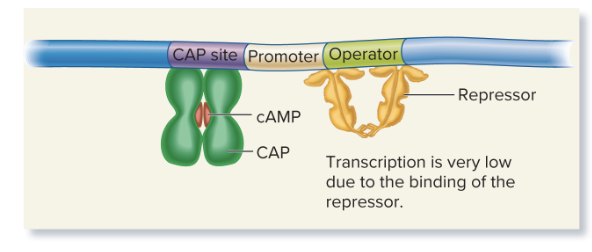
lactose and glucose (low cAMP)
Very low transcription: Low cAMP means CAP doesn’t bind effectively, and even though lactose is present, the repressor may still block transcription.
Glucose levels are a key factor in determining cAMP levels. When glucose is present, adenylyl cyclase (the enzyme that produces cAMP) is inhibited, leading to low cAMP production
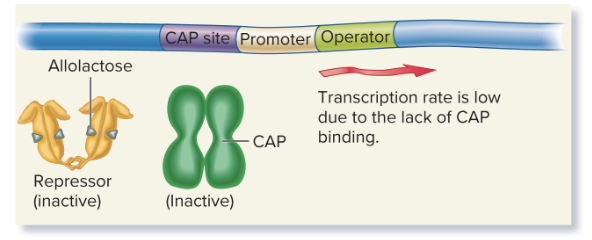
glucose, no lactose (low cAMP)
No transcription: Glucose lowers cAMP, so CAP doesn't bind, and the repressor binds the operator, preventing transcription (of lacZ, lacY, and lacA for lactose metabolization)