CH 16 Molecular Basis of Inheritance.
1/46
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
47 Terms
Explain Griffith’s experiment showing that a genetic trait can be transferred between bacterial strains
experiment demonstrated that a genetic trait can be transferred between bacterial strains by injecting mice with a mixture of live, non-virulent bacteria (R-strain) and heat-killed, virulent bacteria (S-strain), which resulted in the mice developing a deadly infection, proving that the live R-strain had somehow acquired the ability to cause disease from the dead S-strain, a phenomenon he called "transformation" and the transferred substance as the "transforming principle" - later identified as DNA by Avery, MacLeod, and McCarty; essentially showing that genetic material can be transferred between bacteria, allowing for changes in their traits
Define transformation
the process where a cell directly takes up and incorporates exogenous (foreign) genetic material from its surroundings, essentially altering its genetic makeup by integrating this new DNA into its own genome, leading to a stable genetic change within the cell; this is often done in a laboratory setting by artificially inducing a state of competence in the cell to facilitate DNA uptake.
Explain Hersey and Chase’s experiment that showed that DNA is the genetic material
proved that DNA is the genetic material by using bacteriophages (viruses that infect bacteria) labeled with radioactive isotopes to distinguish between the phage's DNA and protein components, demonstrating that only the DNA entered the bacterial cell during infection, thus carrying the genetic information needed to produce new viruses; this confirmed DNA as the genetic material rather than protein, which was previously believed to hold that role.
Predict if radiolabeled DNA (32P) or radiolabeled protein (35S) from phage will infect bacteria
only radiolabeled DNA (32P) from a phage will be able to infect bacteria, as DNA is the genetic material that enters the bacterial cell, while the radiolabeled protein (35S) will remain outside the cell and not be involved in infection.
Explain what Chargaff discovered related to the ratio of adenine and thymine and the ratio of guanine and cytosine
discovered that in DNA, the amount of adenine is always equal to the amount of thymine, and the amount of guanine is always equal to the amount of cytosine, which is known as "Chargaff's rules"; this finding was crucial in understanding the structure of DNA as it revealed the complementary base pairing between these nucleotide pairs.
Given the percentage of a single DNA nucleotide in a genome, predict the percentages of the other three nucleotides
know the percentage of one DNA nucleotide in a genome, you can predict the percentages of the other three nucleotides by understanding base pairing rules: the percentage of adenine (A) will be equal to thymine (T), and the percentage of cytosine (C) will be equal to guanine (G); therefore, to find the remaining nucleotides, simply subtract the known percentage from 100% and divide the remaining percentage equally between the complementary base pair
Explain the technique used by R. Franklin to study DNA
technique called X-ray crystallography to study the structure of DNA, which allowed her to capture images that revealed crucial information about the molecule's helical shape, ultimately contributing significantly to the discovery of the DNA double helix model by Watson and Crick.
Explain what her photo 51 revealed
the double-helix structure of DNA
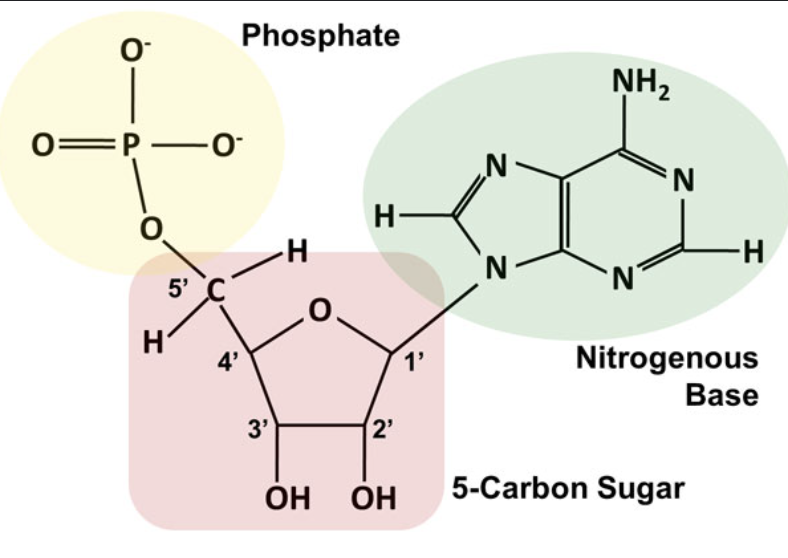
the structure of a DNA nucleotide and label the numbers for the carbons on the deoxyribose sugar. Include the position of the phosphate group and the nitrogenous base and hydroxyl group on the drawing
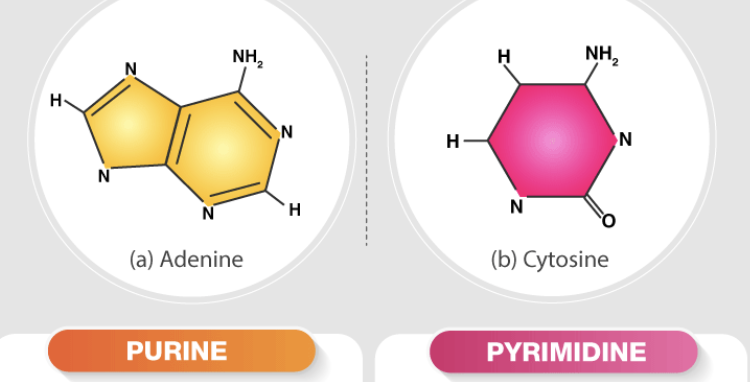
Contrast a pyrimidine vs. purine (structure)
\a single six-membered ring structure containing two nitrogen atoms, while the other one has a larger, double-ringed structure consisting of a six-membered pyrimidine ring fused with a five-membered imidazole ring, containing a total of four nitrogen atoms; essentially, a purine is a pyrimidine ring with an additional ring attached to it, making it larger and with more nitrogen atoms
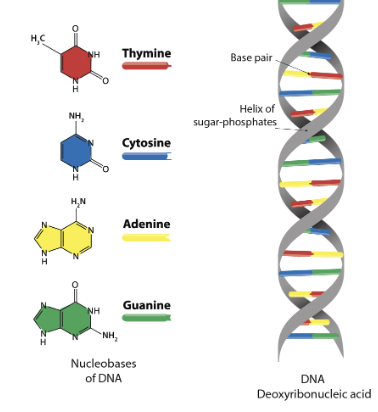
Explain if the helices are parallel or antiparallel and what the directionality means.
In a DNA double helix, the two strands are considered "antiparallel," meaning they run in opposite directions to each other, with one strand's 5' end aligning with the other strand's 3' end; this directionality is crucial for DNA replication and base pairing as it allows for complementary base pairs to form between the strands
Explain where and what the sugar phosphate backbone is and how the bases are linked in the sugar phosphate backbone
the structural framework of a nucleic acid molecule like DNA or RNA, consisting of alternating sugar (deoxyribose in DNA, ribose in RNA) and phosphate groups that are linked together by phosphodiester bonds, forming the outer framework of the molecule, while the nitrogenous bases are attached to the sugar groups within the backbone; essentially, the bases are not directly part of the sugar-phosphate backbone, but rather "hang off" of it, attached to the sugar molecules within the chain
Explain what is meant by complementary base pairing
refers to the specific pairing of nitrogenous bases in DNA, where adenine (A) always pairs with thymine (T) and cytosine (C) always pairs with guanine (G), allowing the two strands of DNA to form a stable double helix structure by connecting with hydrogen bonds; essentially, each base has only one "complementary" partner it can bind with, like a lock and key mechanism, which is crucial for accurate DNA replication and transcription
how the nitrogenous bases are bonded (what TYPE of bond is involved in complementary base pairing
bonded together by hydrogen bonds. This means that the specific pairing between bases like adenine (A) with thymine (T) and cytosine (C) with guanine (G) is maintained by these weak hydrogen bonds
Where are the nitrogenous bases in DNA located?
the nitrogenous bases are located in the center of the double helix, forming the "rungs" of the ladder-like structure between the two sugar-phosphate backbones that run along the outside of the helix; essentially, they are positioned on the inside of the DNA molecule
Explain why complementary base pairing is important for DNA replication
it ensures that when the DNA strands separate during replication, each strand serves as a template for the creation of a new, exact complementary strand, resulting in two identical DNA molecules, each containing one strand from the original DNA and one newly synthesized strand; this process is known as semi-conservative replication and is vital for accurate genetic information transfer during cell division
Explain the three predicted mechanisms for how DNA replicates: conservative, semiconservative, dispersive.
conservative, where the original DNA molecule remains completely intact and serves as a template to create an entirely new double helix
semiconservative, where each strand of the original DNA molecule acts as a template for a new complementary strand, resulting in two new DNA molecules, each containing one old strand and one new strand
dispersive, where the original DNA strands are fragmented and mixed with newly synthesized DNA segments, creating daughter molecules with a patchwork of old and new DNA segments throughout each strand
Define and explain the role of the following enzymes in DNA replication: helicase
"unzipping" the double-stranded DNA molecule, separating the two strands to create a replication fork, which allows other enzymes like DNA polymerase to access each strand and synthesize new complementary DNA strands; essentially, it breaks the hydrogen bonds between base pairs to unwind the DNA helix, making it accessible for replication
Explain where and how DNA nucleotides are added during DNA replication.
new nucleotides are added exclusively to the 3' end of the growing DNA strand by the enzyme DNA polymerase, which "reads" the template strand and adds complementary nucleotides one by one, always moving in the 5' to 3' direction along the template strand; this means that the new strand is synthesized in the 5' to 3' direction as well
Explain 5’ to 3’
refers to the directionality of a nucleic acid strand, like DNA or RNA, where the sequence is read from the end with a phosphate group attached to the 5th carbon of the sugar molecule (the 5' end) towards the end with a hydroxyl group attached to the 3rd carbon of the sugar molecule (the 3' end); essentially, this means new nucleotides are always added to the 3' end during synthesis, making the strand grow in a 5' to 3' direction.
Where does the energy come from for this biosynthesis?
the breakdown of food molecules, which is stored in the form of ATP (adenosine triphosphate), essentially acting as the "energy currency" of the cell; this energy is then used to drive the synthesis of new molecules needed by the organism
What determines WHICH DNA nucleotide is added?
the complementary base pairing with the nucleotide on the template strand; meaning, an "A" will always pair with a "T" and a "C" will always pair with a "G", ensuring the new strand is an exact copy of the original strand.
Define origin of replication and replication fork
a specific DNA sequence on a chromosome where DNA replication begins, essentially the starting point for the copying process, while the other one is the Y-shaped structure that forms at the origin of replication where the DNA strands are actively being separated and copied during replication
Contrast DNA replication on the leading and lagging strand
the leading strand is synthesized continuously in the same direction as the replication fork movement, while the lagging strand is synthesized discontinuously in short fragments called Okazaki fragments, moving in the opposite direction of the replication fork due to the limitations of DNA polymerase which can only add nucleotides in the 5' to 3' direction; this results in the need for multiple RNA primers on the lagging strand to initiate synthesis of each Okazaki fragment, unlike the single primer needed on the leading strand
Define Okazaki fragment
a short segment of DNA that is created during the discontinuous synthesis of the lagging strand during DNA replication
explain the function of DNA polymerase I and ligase
is responsible for removing RNA primers from the newly synthesized DNA strand and replacing them with appropriate DNA nucleotides, while the other one joins the gaps between these newly added DNA fragments (Okazaki fragments) on the lagging strand, creating a continuous DNA molecule
Contrast eukaryotic and bacterial DNA structure, and contrast the numbers of origins of replication
One is linear and has multiple origins of replication along each chromosome, while the other one is typically circular and has only one origin of replication per chromosome; this difference in origin number is primarily due to the much larger size of eukaryotic genomes, requiring replication to start at multiple points to complete duplication in a timely manner
Define mutation, and describe two causes of mutation
a change in the DNA sequence of an organism, which can occur due to errors during DNA replication during cell division or exposure to environmental factors like radiation or chemicals that damage DNA (mutagens)
Explain the process of nucleotide excision repair
DNA repair mechanism that removes bulky lesions, like those caused by UV radiation, from the DNA by recognizing the damage, cutting out a short single-stranded segment containing the lesion, and then replacing it with a new, undamaged piece using the complementary strand as a template, ultimately sealing the gap with DNA ligase
Explain the cause of Xeroderma pigmentosa and its effect
caused by a genetic mutation that impairs the body's ability to repair DNA damage caused by ultraviolet (UV) radiation from sunlight, leading to extreme sensitivity to sun exposure, resulting in skin pigmentation changes, dry skin, and a high risk of developing skin cancers, particularly when exposed to sunlight; essentially, the body cannot effectively fix the DNA damage caused by UV rays, causing the skin to become highly susceptible to mutations that can lead to cancer development
Explain the location, structure, and functions of telomeres
specialized protein-DNA structures located at the very ends of linear chromosomes, acting as protective caps that prevent chromosome degradation and maintain genomic stability by shielding the ends from being mistaken for DNA damage; they consist of repeating DNA sequences, typically "TTAGGG" in humans, bound by specific proteins, and gradually shorten with each cell division, ultimately limiting a cell's replicative lifespan and contributing to aging.
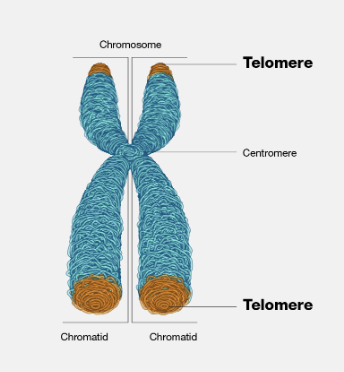
Explain why telomeres shorten and the effects of telomere shortening
because of a biological limitation in DNA replication called the "end replication problem," where the enzymes responsible for copying DNA cannot fully replicate the very ends of chromosomes, resulting in a gradual loss of telomere sequence with each replication cycle; this shortening is associated with aging as once telomeres become critically short, cells enter a state of senescence (permanent cell cycle arrest) and can no longer divide effectively, leading to tissue decline and potential for age-related diseases
Explain the role of telomerase
acts as a "reverse transcriptase" to add repetitive DNA sequences (called telomere repeats) to the ends of chromosomes, essentially maintaining their length and preventing them from shortening with each cell division
Explain WHY DNA is packed within the nucleus
o efficiently organize and protect the long strands of genetic material, allowing for proper cell division, DNA replication, and gene expression by preventing tangling and damage while also enabling controlled access to specific genes when needed; essentially, it acts like a neatly folded map within a designated container, making it readily accessible when required
Define histone
a protein that acts as a spool around which DNA wraps to form compact structures called nucleosomes, essentially packaging DNA into chromatin within the nucleus of a cell; histones play a crucial role in regulating gene expression by allowing access to DNA for transcription factors through various chemical modifications on their protein tails, essentially acting as a key component of epigenetic regulation
Explain how chromatin is packed into a nucleosome, 30 nm fiber, 300 nm fiber, and mitotic chromosome
by wrapping DNA around a histone protein complex, forming a "bead on a string" structure; these nucleosomes then further condense into a 30nm fiber through interactions with linker histones, which then loop and fold to create a 300nm fiber, ultimately leading to the highly condensed mitotic chromosome structure by further compaction and scaffolding proteins during cell division
Explain the experiment used by Meselson and Stahl to discriminate between the possibilities.
Meselson and Stahl used isotope labeling to distinguish between old and new DNA strands. They grew E. coli bacteria in a medium containing "heavy" nitrogen (15N) for several generations, ensuring that all the bacterial DNA was labeled with this heavier isotope
The Meselson-Stahl experiment provided strong evidence for the semiconservative model of DNA replication proposed by Watson and Crick. It demonstrated that during replication, each strand of the original DNA serves as a template for a new complementary strand
Explain which mechanism is, in fact, how DNA replicates.
mechanism called semi-conservative replication; meaning that each strand of the original DNA molecule serves as a template to create a new complementary strand, resulting in two new DNA molecules, each containing one strand from the original molecule and one newly synthesized strand
Define and explain the role of the following enzymes in DNA replication: single-strand DNA binding protein,
a protein that binds to single-stranded DNA during DNA replication, preventing the strands from re-annealing back together and protecting them from degradation by nucleases, essentially allowing other replication enzymes to access and work on the separated DNA strands effectively; it plays a crucial role in stabilizing the single-stranded regions created by DNA helicase at the replication fork
Define and explain the role of the following enzymes in DNA replication: topoisomerase
temporarily breaking and rejoining DNA strands, effectively relieving the tension caused by the unwinding of the double helix at the replication fork, preventing the DNA from becoming excessively twisted or tangled during the process; essentially, it manages the topological state of DNA by introducing transient breaks to allow for strand passage and unwinding, ensuring smooth replication progression
Define and explain the role of the following enzymes in DNA replication: primase
responsible for synthesizing short RNA segments called "primers" which act as starting points for DNA polymerase to begin adding new nucleotides to the DNA strand; essentially, it "primes" the DNA molecule for replication by creating a short RNA sequence that DNA polymerase can then build upon, as DNA polymerases cannot initiate synthesis on a bare DNA strand
Define and explain the role of the following enzymes in DNA replication: DNA polymerase III
the primary enzyme responsible for synthesizing new DNA strands during DNA replication, acting as the main replication enzyme in prokaryotes and playing a crucial role in building both the leading and lagging strands at the replication fork by adding nucleotides in a 5' to 3' direction; it is known for its high processivity, meaning it can add nucleotides to a growing DNA strand very efficiently without detaching frequently
Define and explain the role of the following enzymes in DNA replication: sliding clamp
a ring-shaped protein complex that functions as a key component of the DNA replication machinery by holding the DNA polymerase enzyme tightly onto the DNA template strand, significantly increasing the enzyme's processivity (ability to add nucleotides continuously without detaching) during DNA synthesis; essentially acting like a tether to prevent the polymerase from dissociating from the DNA while it moves along the strand to replicate it
Aneuploidy
Chromosomal aberration in which chromosomes are present in extra copies or are deficient in number
Explain the process of nucleotide excision repair and the role of nuclease
nuclease: to specifically cut the DNA strand on both sides of the lesion, effectively excising the damaged segment so it can be replaced by a new, undamaged piece
Explain the process of nucleotide excision repair and the role of DNA polymerase
This uses the undamaged strand as a template to synthesize a new complementary strand, filling in the gap left by the excised damaged section
Explain the process of nucleotide excision repair and the role of DNA ligase in the repair.
DNA ligase joins the newly synthesized DNA segment to the existing strand by forming a phosphodiester bond, completing the repair process