cell junctions
1/28
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
29 Terms
Gap junctions structure
Connexons/hemichannels - made of connexins
6 connexins make up a connexon
Connexons from neighbouring cells dock to form gap junction (channel opens only when correct docking has occurred)
Gap junctions formation
Connexins synthesised in rER
Connexins oligomerise in Golgi apparatus to form Connexons
Connexons are trafficked to the plasma membrane and inserted near existing junctional complexes (bind to scaffolding protein ZO-1)
Multiple channels cluster to form plaques
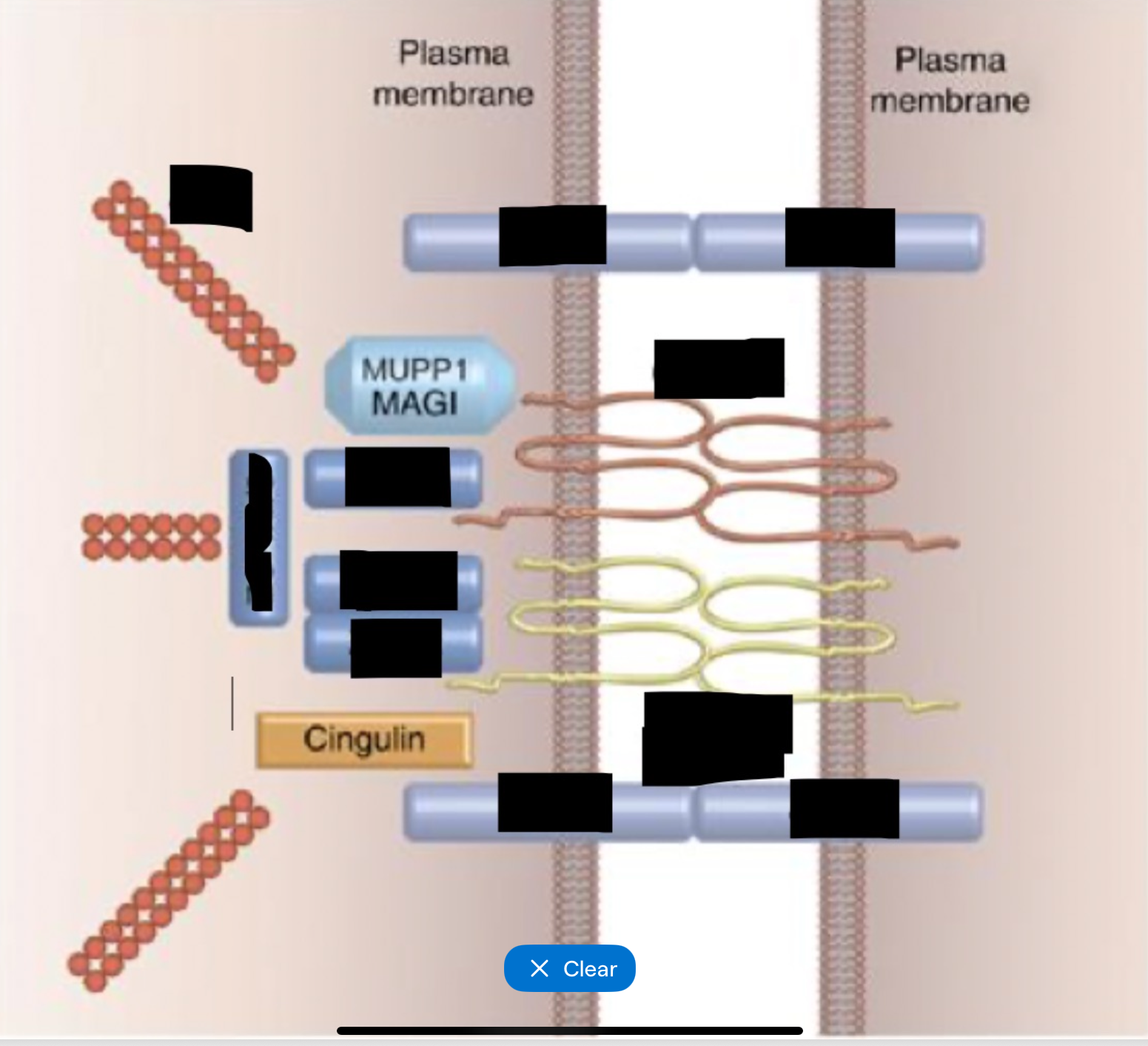
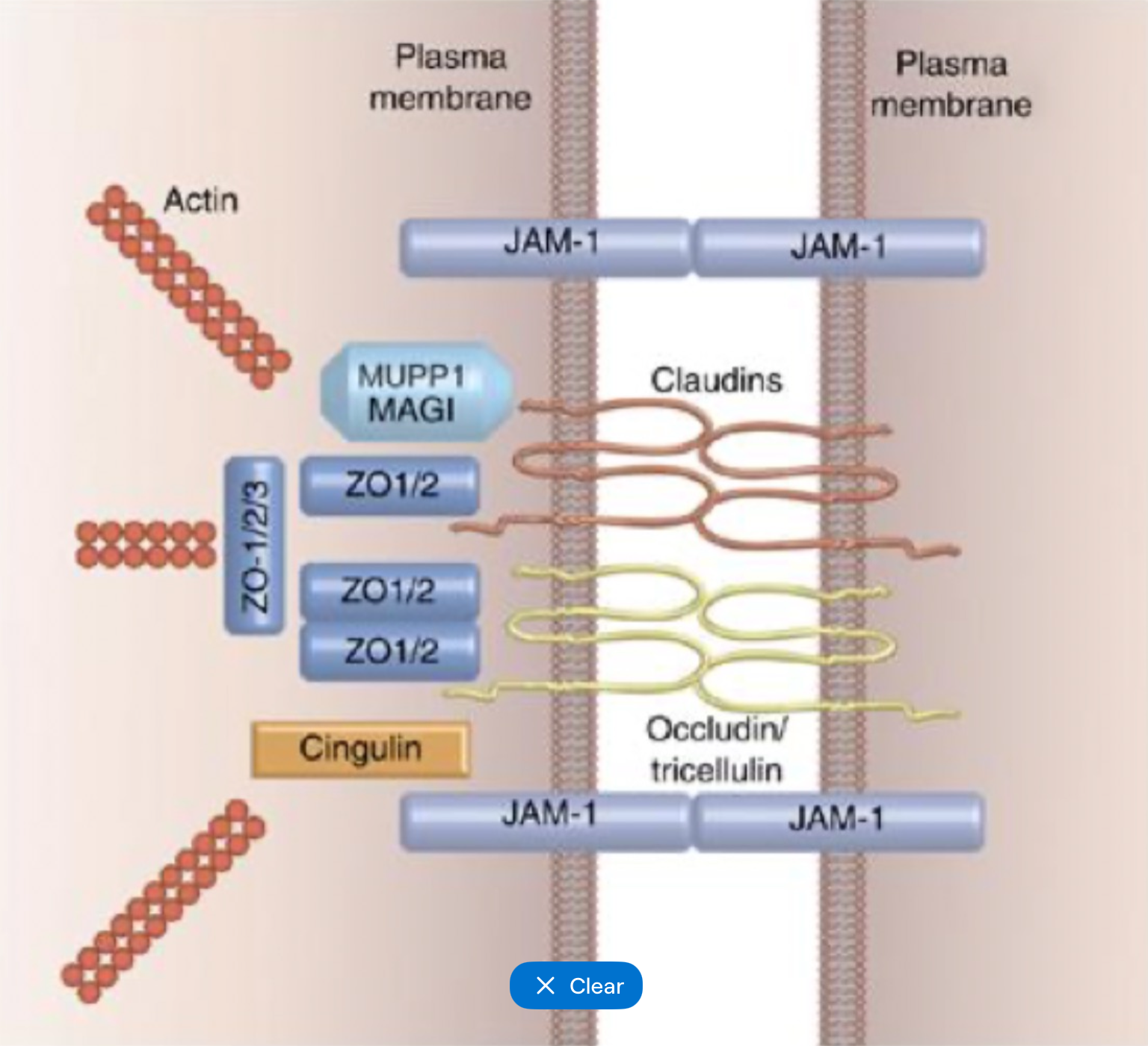
Tight junctions general
Cell-cell adhesion complexes near apical membrane
Selective barrier controls paracellular pathway and maintains cell polarity
Branching network of sealing strands - more strands = more effective at controlling permeability
40+ proteins
Tight junctions transmembrane proteins
Occludin
Junctional adhesion molecules (JAMs)
Claudins
Occludin
Tight junctions
4 transmembrane domains
Important for stability and integrity
Contributes to tightness not permeability
Junctional adhesion molecules (JAMs)
Tight junctions
Transmembrane proteins in Ig superfamily
Mediate cell-cell adhesion
Organise assembly
Interact with cytoskeleton and signalling proteins and scaffolding proteins
Claudins
Tight junctions
Determine permeability
Transmembrane proteins with 4 transmembrane domains Important
Act as selective channels
23 genes for isoforms, each allows or restricts particular ions or molecules - Claudin-1 blocks most ions and water, claudin-2 selective permeability to Na+, K+, Ca2+, water
Tight junction scaffolding proteins
Link tight junctions to actin cytoskeleton
Includes ZO-1, 2 and 3
Tight junctions formation
Formation initiated by adherens junctions
Recruitment of polarity proteins
Claudins and occludins inserted into apical lateral membrane
JAMs dimerise and stabilise cell-cell contact
ZO-1 binds to f-actin
Forms a continuous circumferential belt
Tight junctions dyfunction
Inflammatory bowel disease - reduced claudin 1 and 2, increased claudin 3 = leaky barrier
Loss of claudin 4 and 7 and occludins - linked to invasive and metastatic GI cancers
Claudin 16 and 19 mutations - impaired reabsorption of Mg2+ and Ca2+
Salmonella - effector proteins disrupt tight junctions and allow bacterial invasion
Adherens junctions - general
Belt-like adhesion complexes basal to tight junctions (strong)
Mechanical adhesions - tensile strength
Drives cell shape changes and tissue remodelling
Involved in signalling and establishing cell polarity
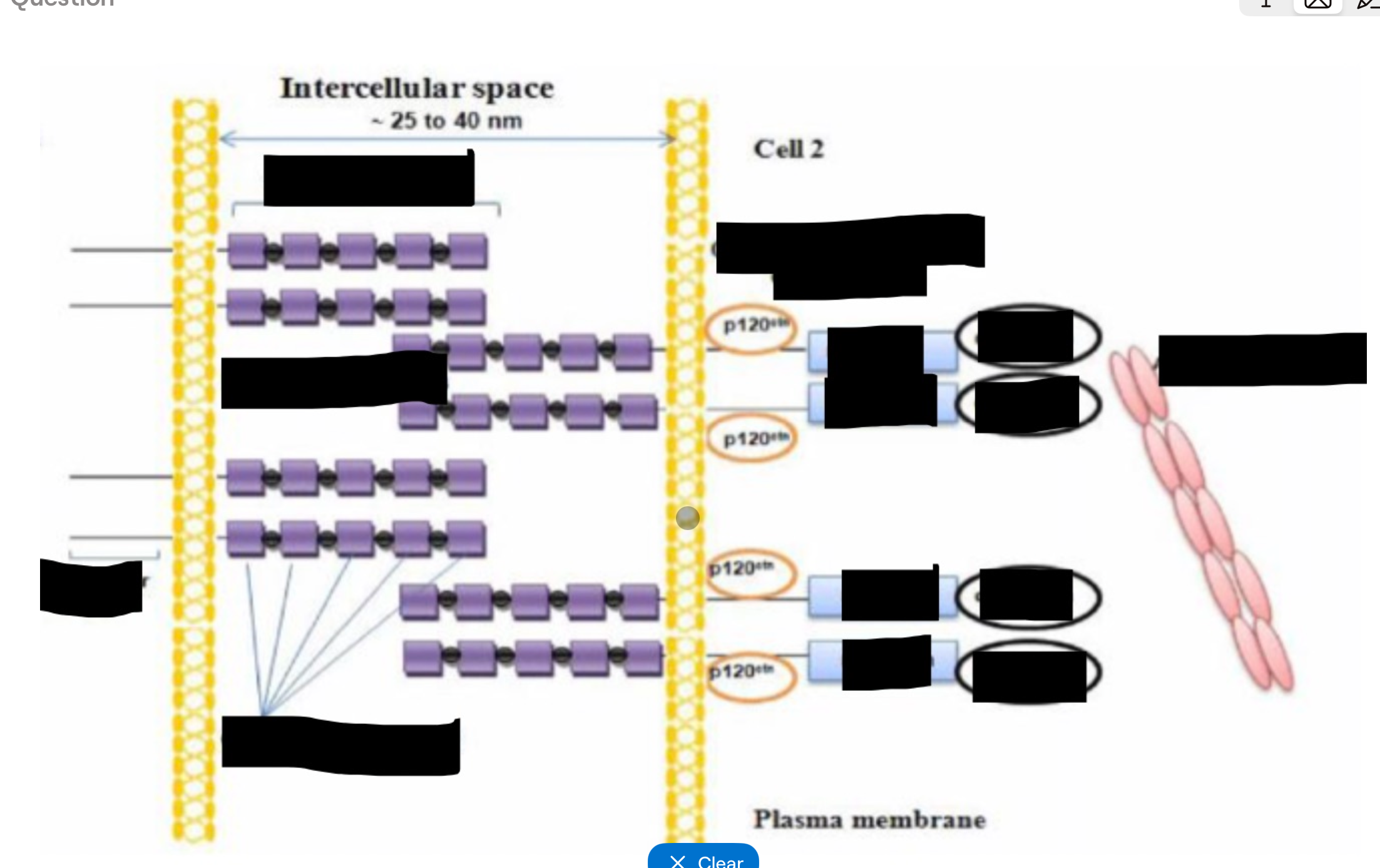
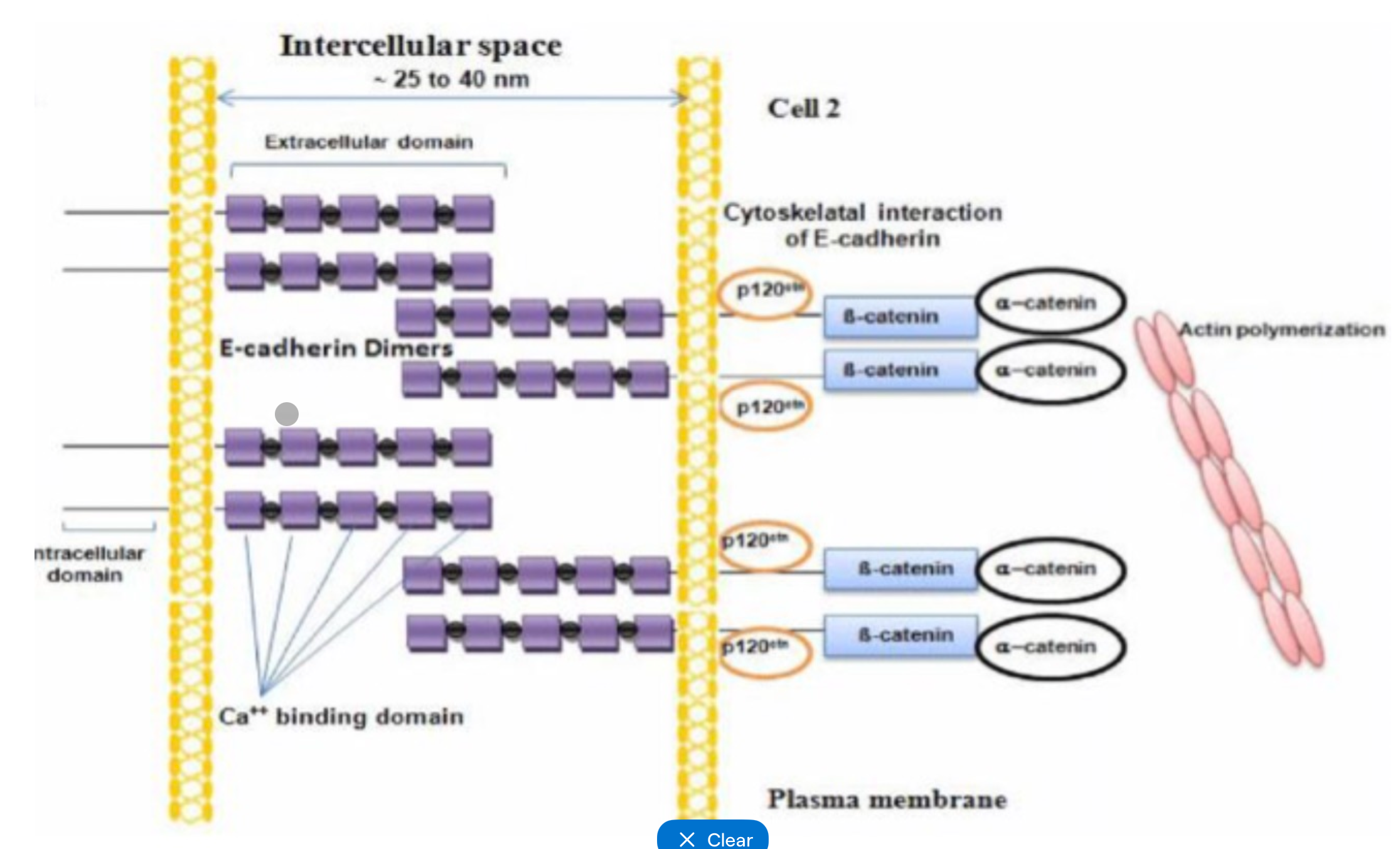
Adherens junctions proteins
Transmembrane = E-cadherins - form dimmers with neighbouring cells dock, calcium dependant binding
Intracellular = catenins - link cadherins to actin cytoskeleton, involved in regulating junction stability and signalling
Adherens junctions formation
Cadherins on adjacent cells form homodimers (calcium dependant)
Cadherin cytoplasmic tails recruit catenins
Catenins connect to actin
Junctions form a continuous circumferential belt
Adherens junctions dysfunction
E-cadherins essential for intestinal epithelial integrity - mutation or loss = increased intestinal permeability
E-cadherins encoded by CDH1 gene - mutations linked to ovarian, breath, thyroid, colorectal and gastric cancers e.g. hereditary diffuse gastric cancers
Desmosomes - general
Mucula adherens
Spot-like intercellular junctions
Link to cytokeratin
Strong mechanical adhesion
Abundant where high mechanical/shearing force
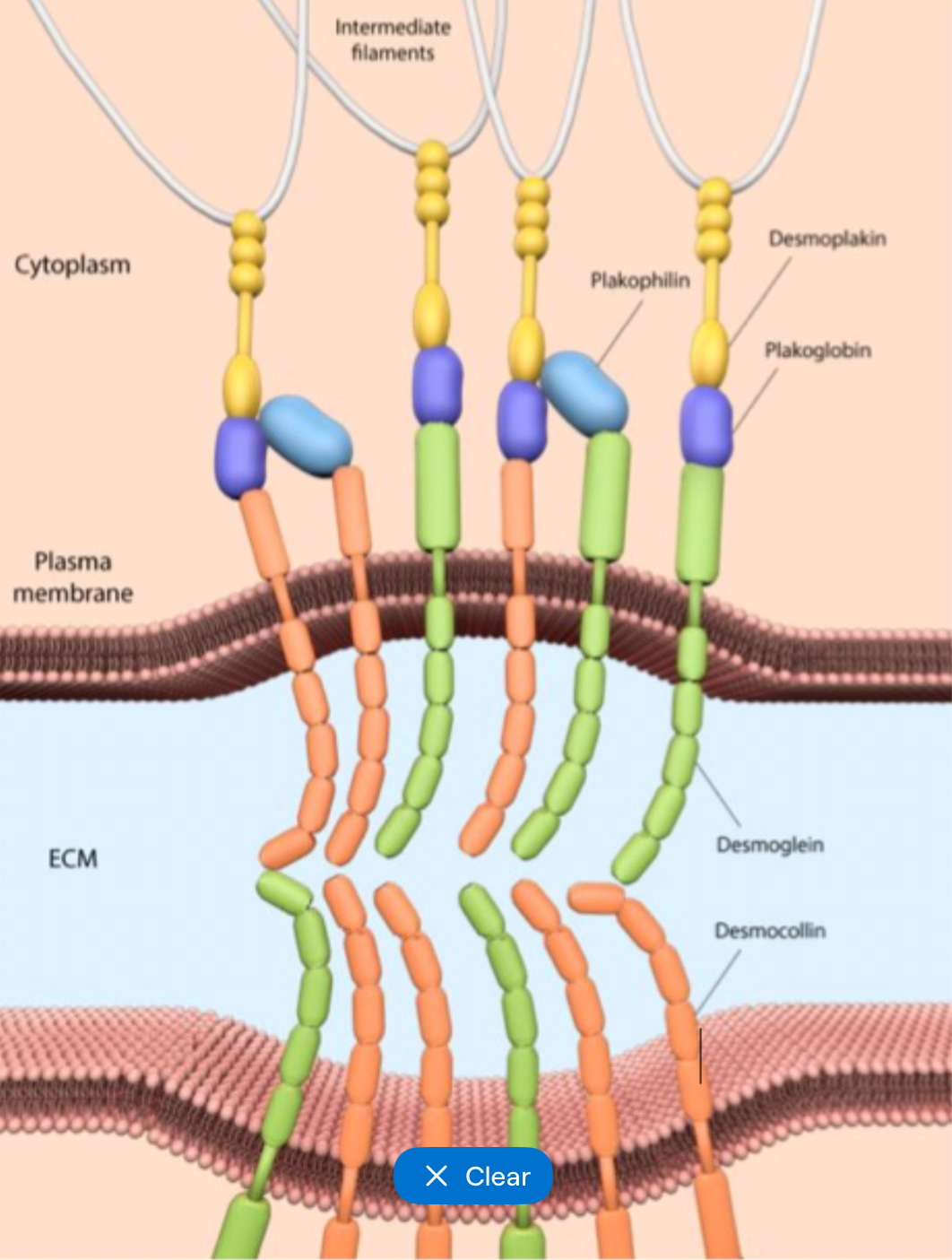
Desmosomes general structure
Desmosomal intermediate filament complex (DIFC)
Transmembrane proteins
Intracellular proteins
Desmosomal intermediate filament complex
Extracellular core with dense plaques
Desmosomes transmembrane proteins
Desmosomal cadherins
Desmogleins and desmocollins
Desmosomes Intracellular proteins
Plakoglobin and desmoplakin
Connect transmembrane proteins to intermediate filaments
Desmosomes formation
Desmogleins and desmocollins on adjacent cells bind
Cytoplasmic tails recruit plakoglobin
Plakoglobin binds to desmoplakin
Desmoplakin connects to cytokeratin
hemidesmosomes - general
Anchor epithelia to basement membrane
Transmembrane protein is integrity
Intracellular protein is pectin
Extracellular protein is laminin (lamina Lucida)
Hemidesmosomes formation
Integrin heterodimers inserted into basal membrane
Binds laminin - binds to basal lamina components
Integrin binds to plectin - links Integrin to keratin
Hemidesmosomes cluster to form dense adhesion sites
Focal adhesions general
Highly dynamic multi protein complexes
Mediate attachment of cell to ECM, connect to actin
More plastic than Hemidesmosomes
Involved in cell signalling and migration
Focal adhesions structure and formation
Transmembrane protein - integrins
Adaptor proteins - e.g. talins
Focal adhesion kinase (FAK) regulates adhesion turnover
Focal adhesions formation
Integrins bind to ECM components (fibronectin, collagen)
Recruitment of adaptor proteins
Link to actin
Cell junction complex formation
Early assembly required for correct positioning and development of apical polarity complexes
1. Initial cell-cell contact
2. Adherens junctions assemble (E cadherins clustering, actin remodelling, polarity initiated)
3. Tight junction formation (recruitment and positioning of polarity determining complexes (act as molecular landmarks, includes recruitment of ZO-1)
4. Desmosome formation
5. Hemidesmosomes and focal adhesions form
Gap junctions form after adherens and tight junctons, as needed