Lecture 20: CRISPR-Cas9
1/16
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
17 Terms
Streptococcus pyogenes
Type of Group A streptococcal (GAS) bacteria that is the causative agent of strep throat
Bacteriophage
An organism that is neither prokaryotic nor eukaryotic and has an outer protein coat surrounding its DNA genome
What does CRISPR stand for?
Clustered Regularly Interspaced Short Palindromic Repeats array
Cas9
RNA-guided endonuclease that catalyzes site-specific cleavage of double-stranded DNA
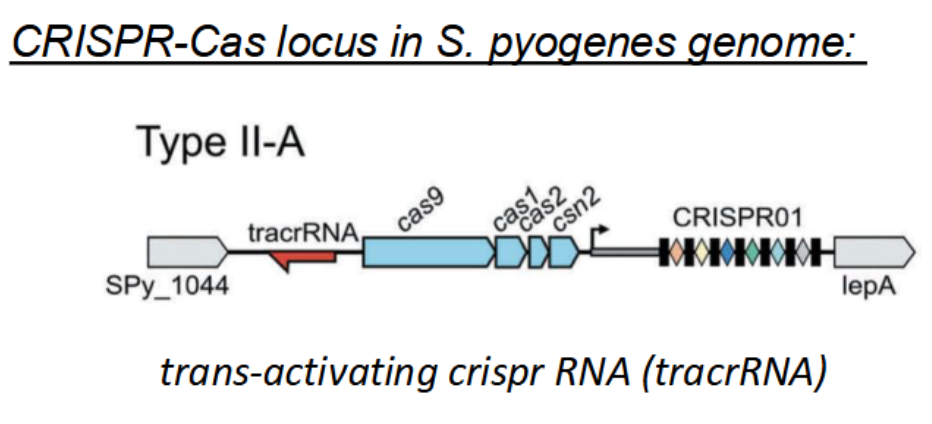
Describe how the CRISPR-Cas9 system works in bacteria.
Adaptation — Insertion of a new spacer, derived from phage DNA into the CRISPR DNA array
CRISPR RNA biogenesis — Transcription and processing of mature CRISPR RNA (crRNA)
Interference — Recognition, cleavage, and destruction of phage DNA via the crRNA/Cas protein complex
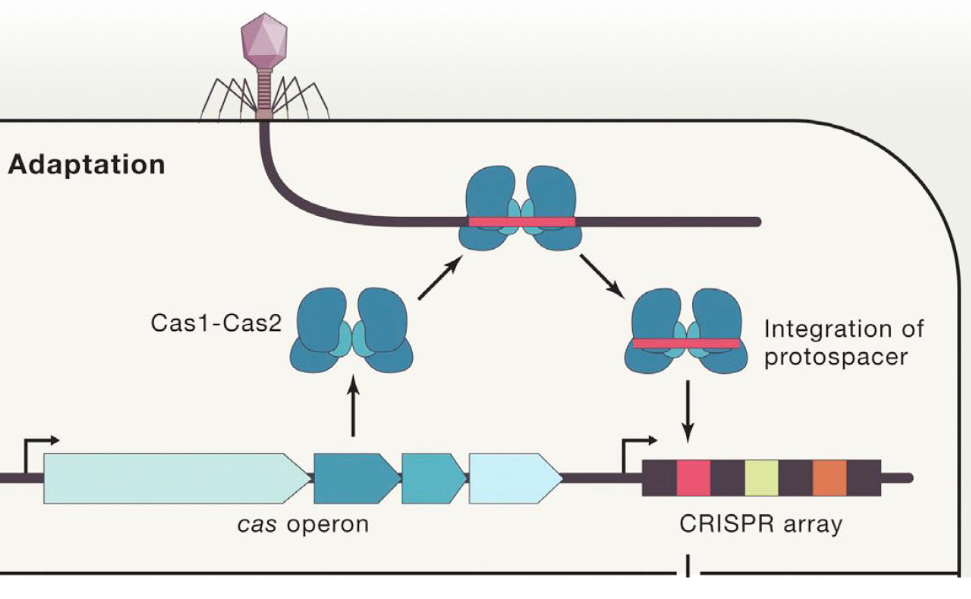
Adaptation
The first step in the CRISPR-Cas9 bacterial immune system response against viruses
Virus injects viral DNA into the bacterium
Bacterial Cas1 and Cas2 enzymes cleave a segment of the viral DNA (protospacer) and integrate the protospacer into the CRISPR DNA array in the bacterial genome
CRISPR RNA biogenesis
The second step in the CRISPR-Cas9 bacterial immune response against viral infections
The CRISPR DNA array is transcribed to make a long CRISPR RNA (crRNA) molecule
A small hairpin trans-activating CRISPR RNA (tracrRNA) binds to a segment of the crRNA, which signals Cas9 to bind to form a complex
RNase III enzyme cleaves the crRNA where the tracrRNA and Cas9 have formed a complex, forming the CRISPR-Cas9 complex → includes Cas9, crRNA, and tracrRNA
crRNA and tracrRNA are types of non-coding RNA molecules

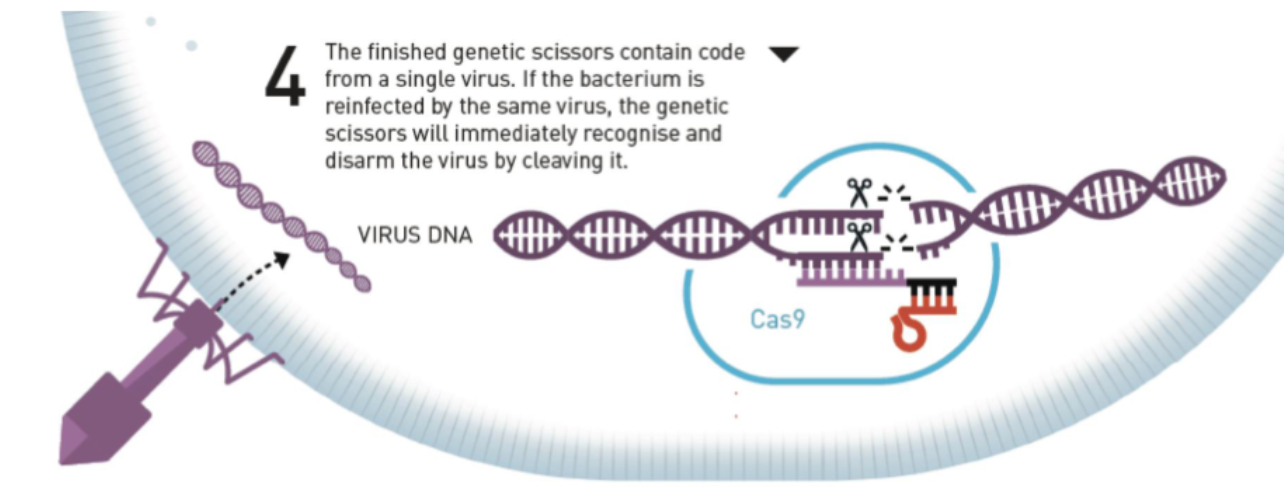
Interference
The third step in the CRISPR-Cas9 bacterial immune response against viral infections
The assembled CRISPR-Cas9 complex contains crRNA from a single virus
crRNA acts as the guide for the Cas9 enzyme to locate the correct bacterial DNA sequence
tracrRNA acts as scaffolding for the complex
S. pyogenes Cas9 recognizes the PAM site NGG, and cleaves between the third and fourth nucleotides 5’ PAM site
If the bacterium is reinfected by the same virus, the CRISPR-Cas9 complex will recognize and “disarm” the virus by cleaving its DNA at the same site
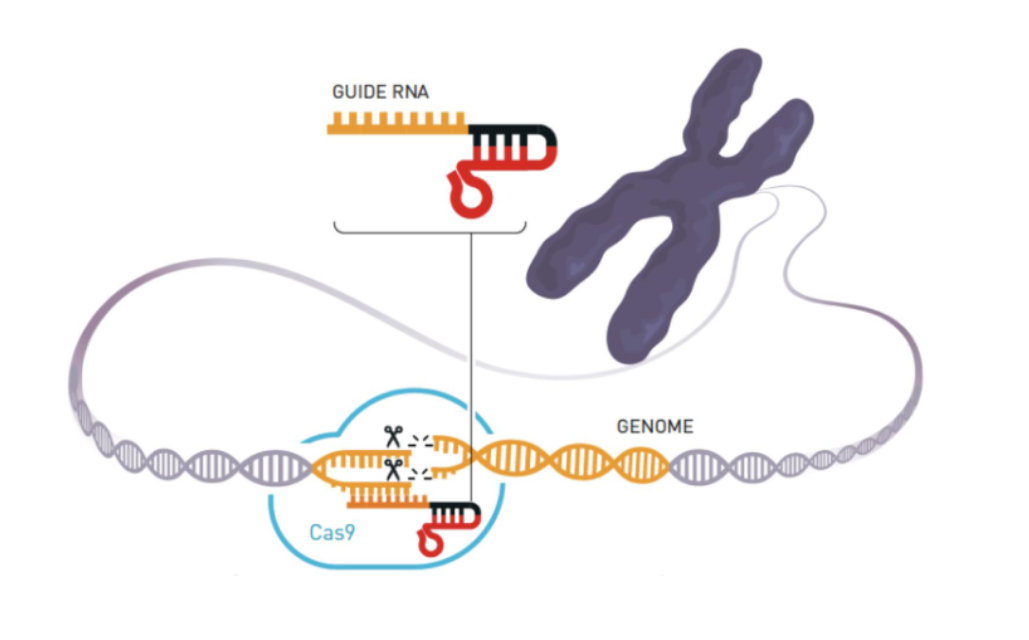
Describe how the CRISPR-Cas9 bacterial immune defense system can be reprogrammed for biotechnology. Natural CRISPR-Cas9 recognizes DNA from bacteriophages, but can it cut any DNA molecule at a predetermined site?
Researchers fused tracrRNA and crRNA into a single molecule called guide RNA (gRNA) → gRNA was the key to modifying the CRISPR-Cas9 system so that it cuts the DNA at a location decided by the researcher
Cas9 enzyme forms a complex with gRNA
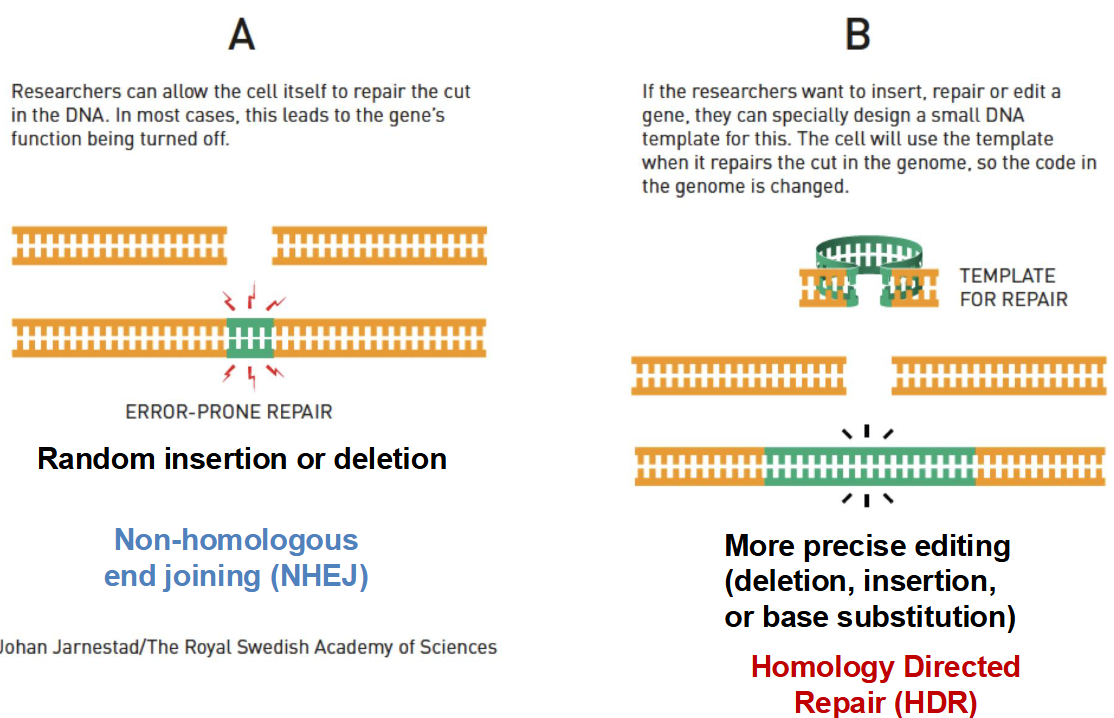
Describe how double-stranded DNA breaks are repaired.
1) Non-homologous end joining (NHEJ) — if the cell itself is allowed to repair the break in DNA with random nucleotides, it usually results in a random insertion or deletion mutation that inactivates the gene (error-prone because the cell does not have a DNA template, occurs during G1)
2) Homology-directed repair (HDR) — if the researchers insert a target DNA sequence to be incorporated, the cell will use the template when it repairs the cut in the genome (accurate and allows for more precise gene editing, occurs during S)
Bacteria acquire immunity to bacteriophages through CRISPR adaptation, biogenesis, and interference. Which of the following is not a required step or component of the bacterial CRISPR system?
A) Assembly of the Cas9-guide RNA complex
B) Insertion of a new phage DNA sequence into the CRISPR cassette
C) tracrRNA attaches to crRNA spacer
D) Processing of mature CRISPR RNA by RNase III
E) None of the above
Answer: E) None of the above
Which of the following is a required component for gene editing by the CRISPR-Cas9 system?
A) CRISPR RNA
B) tracrRNA
C) guide RNA
D) siRNA
E) All of the above
Answer: C) guide RNA
guide RNA includes crRNA and tracrRNA, and in the context of gene editing in the lab, guide RNA is necessary to cause an insertion, deletion, or substitution of the gene of interest
crRNA acts as a template and tracrRNA acts as a scaffold, so gRNA (combination of the two) is required for gene editing
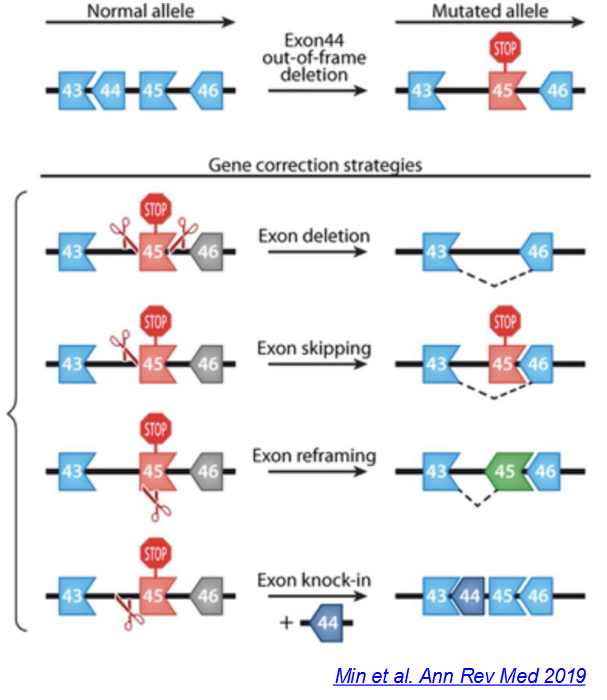
Describe how CRISPR-Cas9 can be used to treat Duchenne muscular dystrophy.
Duchenne muscular dystrophy
Caused by mutations in the X-linked dystrophin gene, disproportionately affecting 1 in 5000 boys worldwide
The allele for dystrophin (43-44-45-46) has an exon 44 frameshift deletion that results in a mutated allele with exon 44 deleted and a nonsense mutation in exon 45 (43-__-45-46)
Gene correction strategies:
Deletion of exon 45 (43-__-__-46)
Skipping of exon 45 (43-__-
45-46)Reframing of exon 45 (43-__-45-46)
Knock-in of exon 44 (43-44-45-46)
nCas9
An endonuclease that has been modified so that it lacks endonuclease ability and cannot cut DNA
Fuses to cytidine deaminase (causes a transition mutation) or adenine deaminase (causes a transversion mutation)
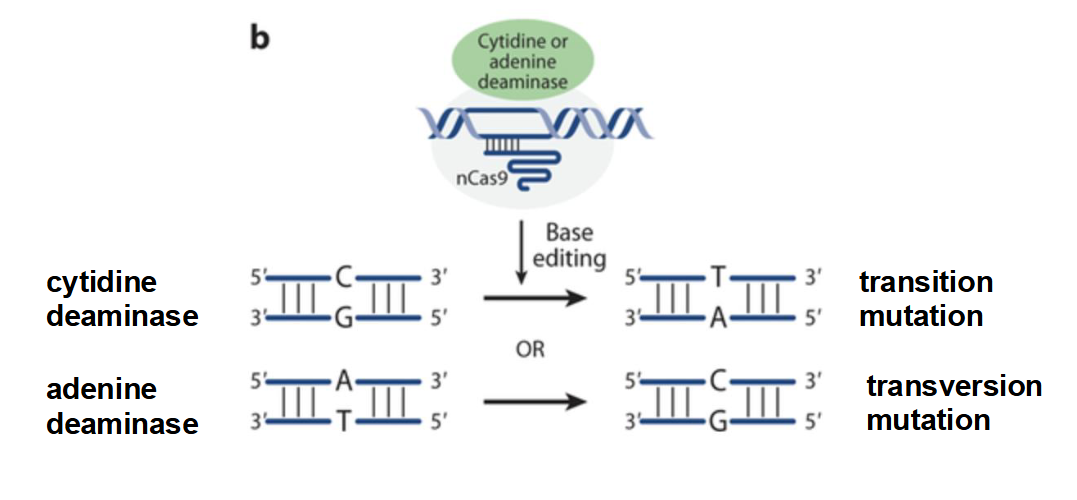
Transition mutation
purine → purine (A→G, G→A)
pyrimidine → pyrimidine (T→C, C→T)
Transversion mutation
purine → pyrimidine (A→C, G→T)
pyrimidine → purine (C→G, T→A)
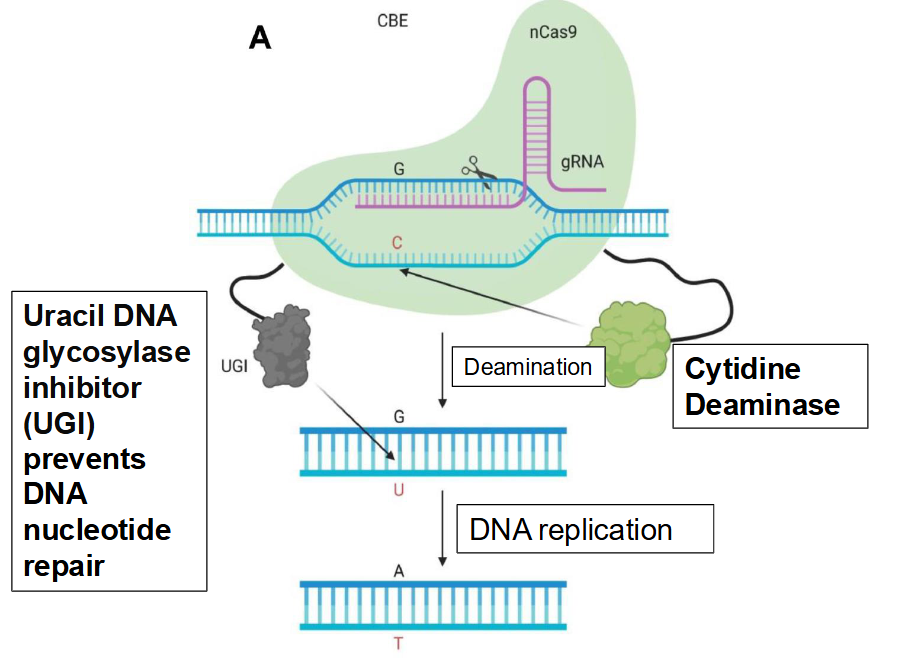
Describe the mechanism of nCas9-deaminase base substitution.
nCas9 binds to either cytidine deaminase or adenine deaminase → instead of making a dsDNA break, the complex makes a ssDNA base substitution
Cytidine deaminase generates transition mutations
Adenine deaminase generates transversion mutations
Ex. nCas9-cytidine deaminase complex localizes to a segment of the DNA sequence
nCas9 opens up the dsDNA where there is a G and C
Cytidine deaminase deaminates G→A and C→U, and uracil DNA glycosylase inhibitor (UGI) prevents DNA nucleotide repair by converting U→T
DNA is replicated as A and T
Overall change: G-C to A-T