Evolution of Populations - Lecture Notes Review (Video)
1/49
Earn XP
Description and Tags
This set of flashcards covers key concepts from the Evolution of Populations lecture notes, including mutation, variation, natural selection, genetic drift, gene flow, founder/bottleneck effects, and Hardy-Weinberg genetics.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
50 Terms
What is the sources of genetic variation in populations?
Mutations
Altering gene number or position
Sexual Reproduction
Which mutations can be passed to offspring in most multicellular organisms (excluding plants)?
Germline mutations - Mutations in cells that produce gametes (sperm & egg).
Why are many mutations harmful in organisms adapted to their environment?
Many mutations change protein structure and/or function and are selected against; in diploids, recessive harmful mutations can be hidden in heterozygotes.
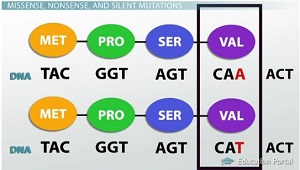
Define a silent mutation.
A base change that does not alter the amino acid sequence of a protein; type of point mutation
What are mutations in non-coding regions typically called?
Neutral variation.
What is a chromosomal mutation?
Mutations that delete, disrupt, or rearrange many loci; often harmful.
What is the significance of gene duplications?
Duplications can increase genome size and provide raw material for new gene functions over time.
Give an example of a gene copy expansion mentioned in the notes.
An ancestral odor-detecting gene has been duplicated many times in humans (about 350 copies) and mice (about 1,000 copies).
How do mutation rates compare among animals/plants, prokaryotes, and viruses?
Animals/plants: about one mutation per 100,000 genes per generation; prokaryotes generally have lower per-generation rates but accumulate mutations quickly due to short generation times; viruses have high mutation rates due to rapid generations.
What role does sexual reproduction play in genetic variation?
It shuffles existing alleles into new combinations via crossing-over, independent assortment, and fertilization.
What is microevolution?
A change in allele frequencies in a population over generations.
Name the three mechanisms that cause allele frequency change.
Natural selection, genetic drift, and gene flow.
Which mechanism is responsible for adaptive evolution?
Natural selection.
How does natural selection alter allele frequencies?
Differential reproductive success causes certain alleles to be passed to the next generation at higher frequencies.
What are the three modes of natural selection?
Directional selection, disruptive selection, and stabilizing selection.
What does directional selection do to trait distributions?
Shifts the mean in one direction, favoring one extreme.
What does disruptive selection do to trait distributions?
Favours extreme phenotypes at both ends, reducing mid-range phenotypes.
What does stabilizing selection do to trait distributions?
Favors intermediate phenotypes and reduces variation.
What is sexual selection and what can it lead to?
A form of natural selection where certain inherited traits increase mating success, potentially leading to sexual dimorphism.
What are the two types of sexual selection?
Intrasexual selection (competition within a sex for mates) and intersexual selection (mate choice by one sex).
What is the 'good genes' hypothesis?
If a trait indicates genetic quality or health, both the trait and the preference for it increase in frequency.
Define genetic drift.
Random fluctuations in allele frequencies, which are more pronounced in small populations and can reduce genetic variation.
What is the founder effect?
A small number of individuals establish a new population with a gene pool that may differ from the original population.
What is the bottleneck effect?
A sudden reduction in population size that may reduce genetic variation and alter allele frequencies.
Give an example of a population affected by the founder effect or bottleneck.
Examples include Tristan da Cunha (founder effect), Polynesian field crickets, Pingelap island (bottleneck), and greater prairie chickens (habitat loss).
Why is genetic drift especially impactful in small populations?
Because random changes in allele frequencies can become fixed more easily when there are few individuals.
What is gene flow?
The movement of alleles among populations, often via migration or dispersal of individuals or gametes.
How does gene flow affect genetic variation among populations?
It tends to reduce differences between populations by homogenizing allele frequencies.
Can gene flow ever decrease a population's fitness?
Yes; if immigration introduces alleles that are maladaptive to local conditions, reducing local adaptation.
Can gene flow ever increase a population's fitness?
Yes; for example, spread of resistance alleles (e.g., insecticide resistance) across populations can increase fitness.
What is the Hardy-Weinberg principle?
A null model describing the genetic makeup of a non-evolving population at a given locus.
List the five conditions for Hardy-Weinberg equilibrium.
No mutations, random mating, no natural selection, extremely large population size, no gene flow.
How do you determine if a real population is evolving using Hardy-Weinberg?
Compare observed allele/genotype frequencies to HW expectations; deviations suggest evolution.
In a population with two alleles, what are p and q?
p is the frequency of the dominant allele, q is the frequency of the recessive allele, with p + q = 1.
What is the HW equation relating genotype frequencies to allele frequencies?
p^2 + 2pq + q^2 = 1, where p^2 is homozygous dominant, 2pq is heterozygous, and q^2 is homozygous recessive.
If PKU occurs in 1 in 10,000 births, what are q^2 and q?
q^2 = 0.0001; q = 0.01; p = 0.99.
Given q = 0.01, what is the carrier frequency (2pq)?
Approximately 2 × 0.99 × 0.01 ≈ 0.0198 (about 1.98%).
In the soybean leaf-color study, what is the procedure to test for evolution using HW?
1) Calculate allele frequencies (p and q). 2) Calculate expected genotype frequencies with HW. 3) Determine observed genotype frequencies. 4) Compare expected vs observed to assess evolution.
In the two-allele flower example (CR and CW), what are p, q, and the expected genotype frequencies?
p = 0.8, q = 0.2; p^2 = 0.64 (CRCR), 2pq = 0.32 (CRCW), q^2 = 0.04 (CWCW).
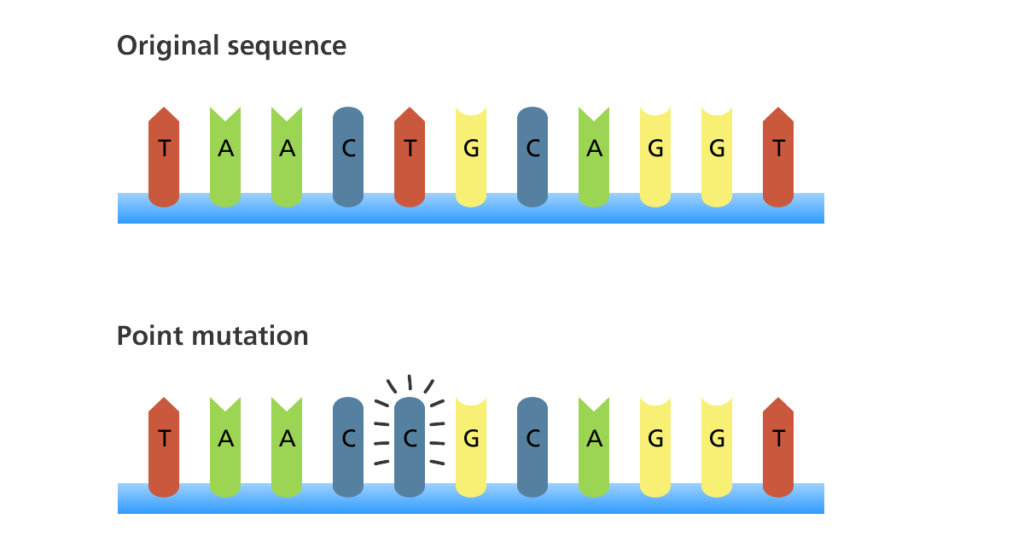
Point Mutation
Change in a single base pair; effect can vary