gaussian discriminant analysis
1/8
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
9 Terms
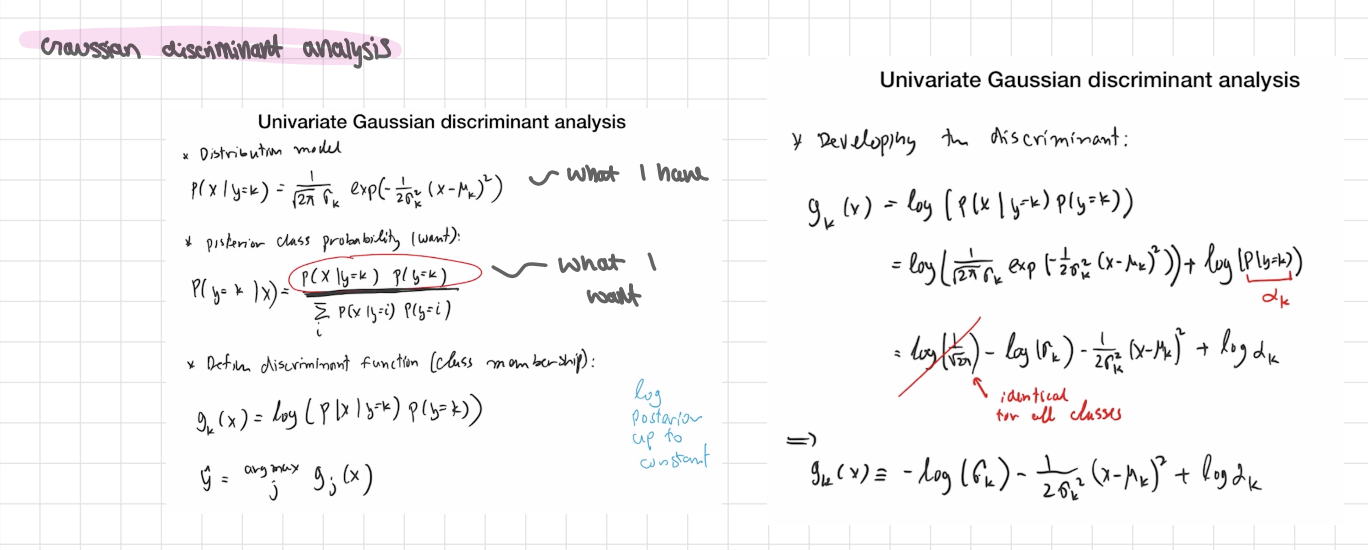
univariate gaussian discriminant analysis
Setup (per class k)
Assume the feature x inside class k is Gaussian:
P(x∣y=k) = N (x;μk,σk²).
Prior of class k: πk=P(y=k)
What we want for prediction. - posterior class
P(y=k∣x) By Bayes:
P(y=k∣x) ∝ P(x∣y=k)πk
For choosing the class, the denominator cancels—so just compare the scores
scorek(x)= P(x∣y=k) πk
Log-discriminant (same thing, easier math)
gk(x) =logP (x∣y=k)+logπk
Plug the Gaussian in and drop the constant −1/2 log(2π) (it’s identical for all classes):
gk(x) = − log σk − (x − μk )² / 2 σk²m + log π k
Predict:
ŷ = =argkmax gk(x)


simplifications
GDA collapses to super simple rules under common assumptions
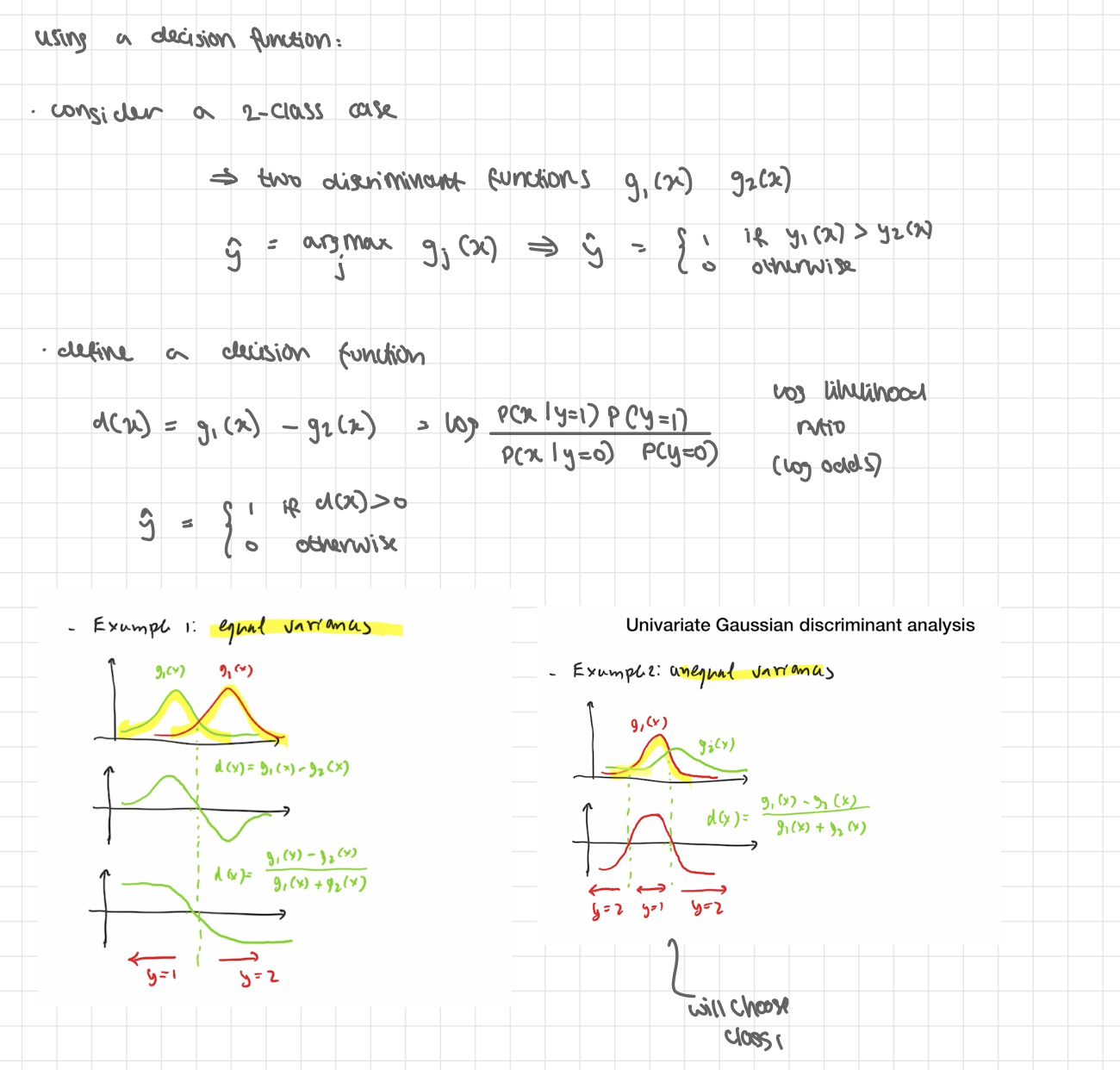
Simplification 1: Equal variances (σk=σ for all k)
The −logσk term is the same for every class, so it doesn’t affect which is largest
→ gk(x) = - 1/2σ² (x- μk )² + logπk
Simplification 2: Equal priors as well (πk same for all k)
Now logπk is also common and drops out:
→ - (x- μk )² → choose class w closest mean

univariate gaussian discriminant analysis summary
gk(x) measures distance to mean. it uses the Mahalanobis distance if variances Arne’t equal. Bias is applied using class priors (higher bias for more likely classes)
process:
1) choose distribution model (Gaussian)
2) compute distribution parameters from data (σk, μk , πk )
3) compute discriminant function gk(x)
4) choose ŷ = argmax j gj(x)

using a decision function
consider a 2-class case
→ 2 discriminant functions g 1(x) g2(x)
ŷ = argmax j gj(x) → ŷ = 1 if y1(x) > y2(x) , 0 otherwise
define a decision function
d(x) = g1(x) - g2(x) = log (P(x | y=1)P(y=1) / P(x|y=0)P(y=0) ) log odds
ŷ = 1 if d(x) >0, 0 otherwise
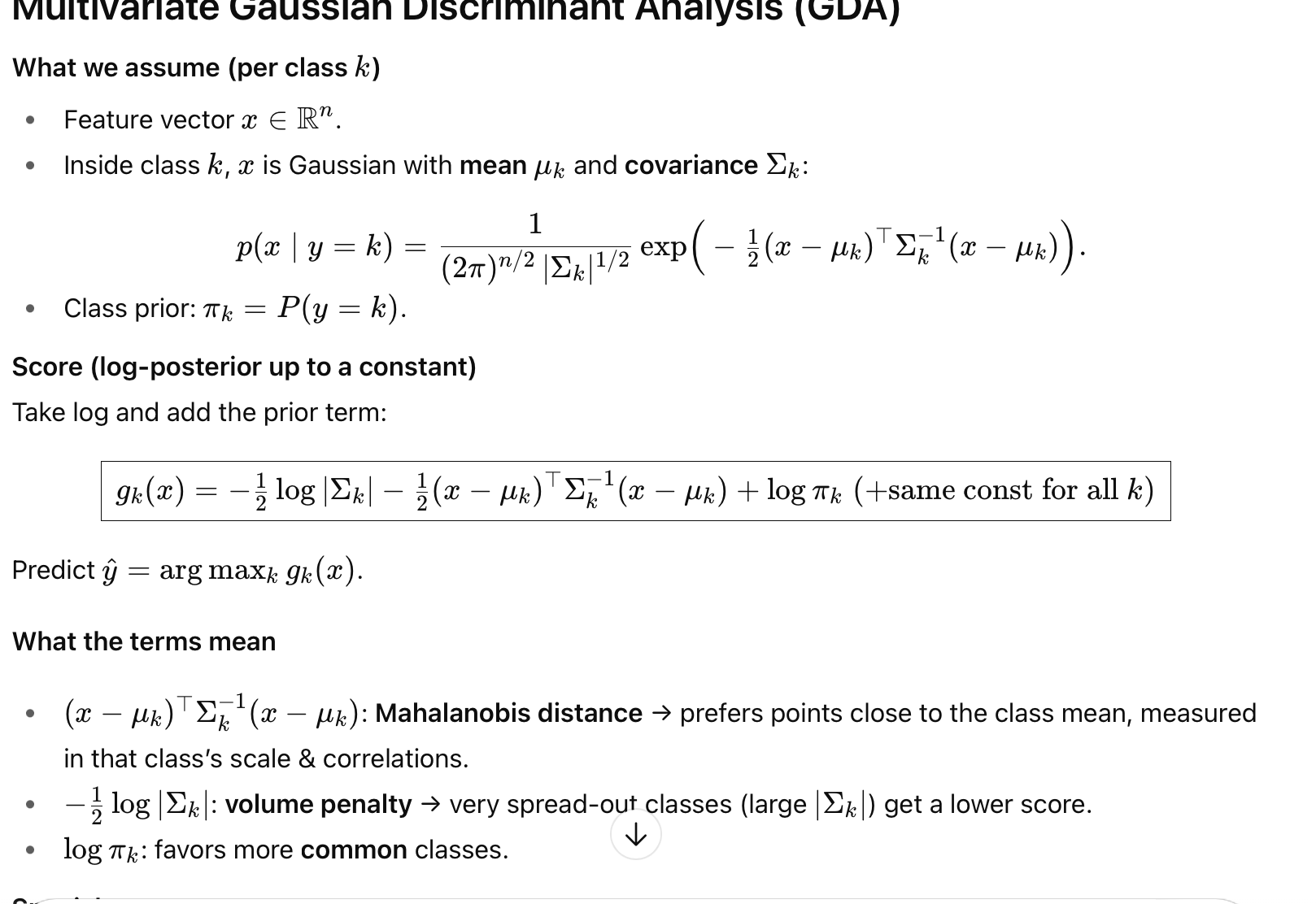
multivariate gaussian discriminant analysis
{ (x(i), y(i))}
What we assume (per class k)
Feature vector x ∈ R n
Inside class k, x is Gaussian with mean μ k and covariance Σk :
(check pic)
Class prior: π k = P ( y = k )
log-posterior up to a constant:
Take log and add the prior term:
gk( x ) = −1/2 log∣Σk∣ − 1/2 (x−μk)⊤ Σk −1 (x−μk)+logπk (+same const for all k)
then predict the label with the biggest gk(x)
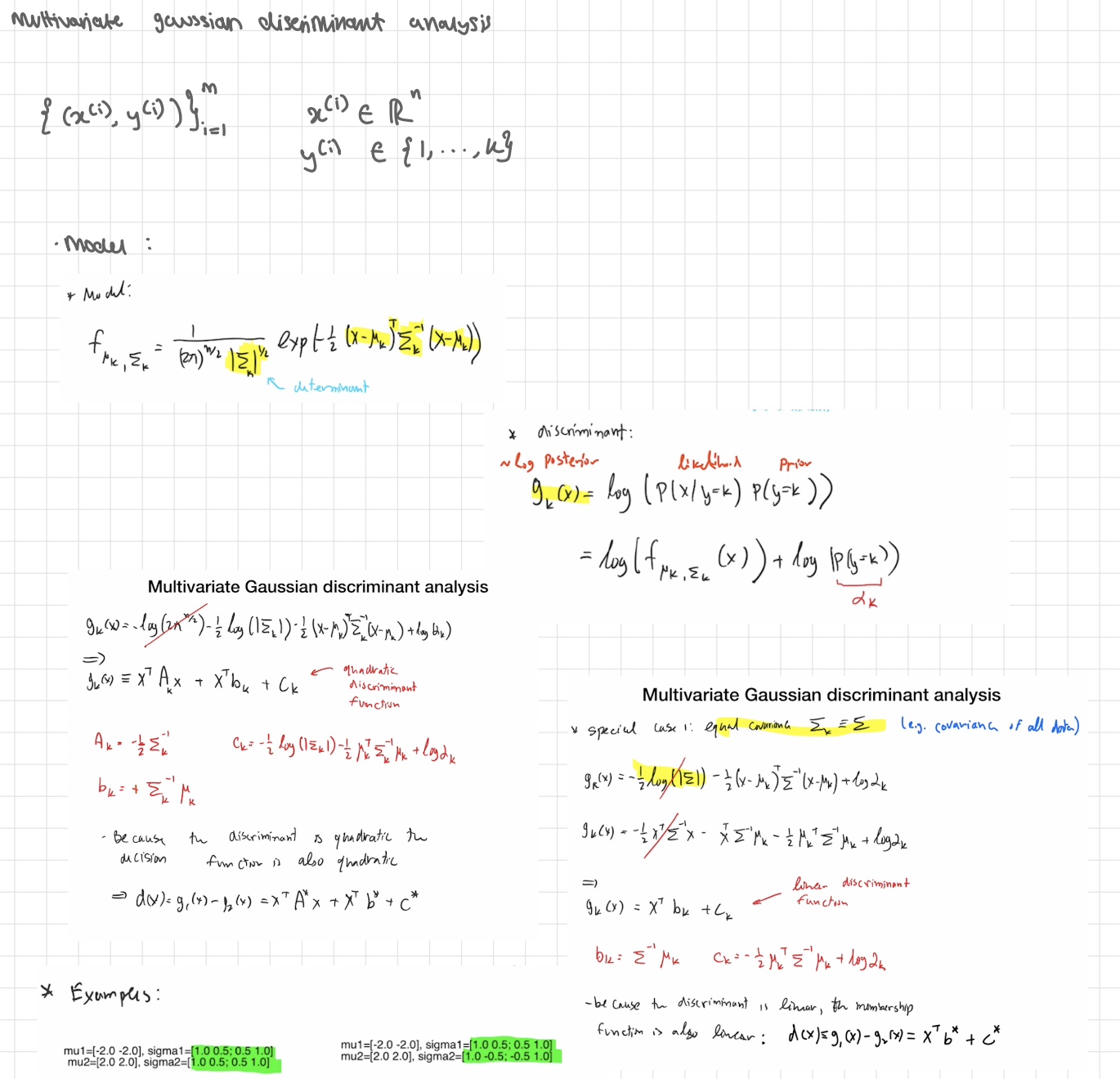
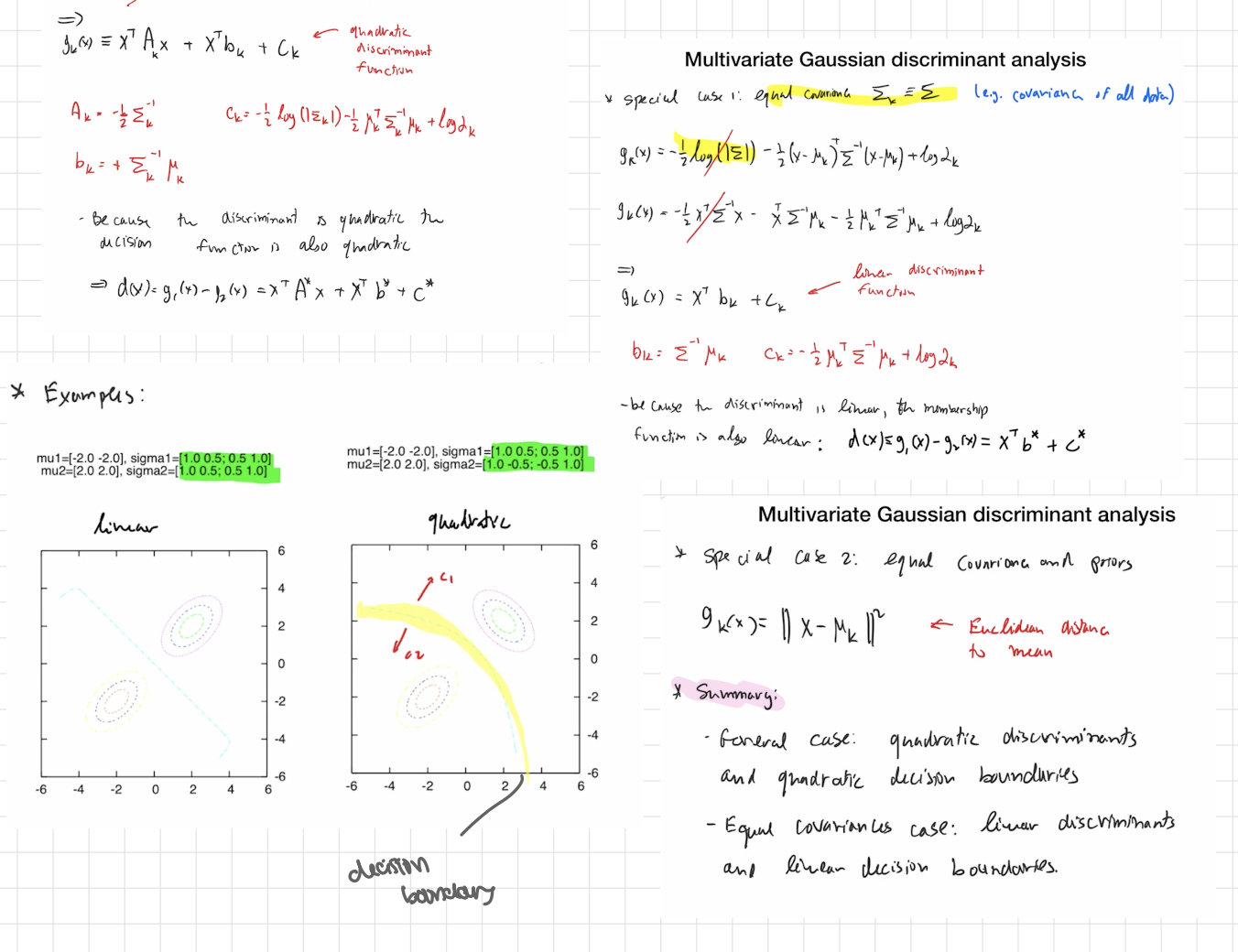
cases
General case ⇒ quadratic rule (QDA)
Expand the square:
gk(x) =x⊤Akx + x⊤bk+ck
with (see pics)
The difference between two classes a,b:
dab(x)=ga(x)−gb(x)= x⊤A*x+x⊤b+ c*
Because of the x⊤(⋅)x term, the boundary dab(x)=0 is quadratic (curved).
That’s why your left plot says “quadratic” and shows a bent boundary.
Special case: equal covariances (Σk=Σ for all k) ⇒ linear rule (LDA)
The quadratic terms cancel (Aa=Ab), so
gk(x)=x⊤bk+ck,
bk=Σ−1μk, ck=−1/2 μk⊤Σ−1μk+logπk
Decision boundary between two classes is a hyperplane (a straight line in 2D)
special case 2: equal covariances and equal priors
Then logπk is the same for all k and drops out.
gk(x)= || x - μk || ² ← euclidean distance to mean
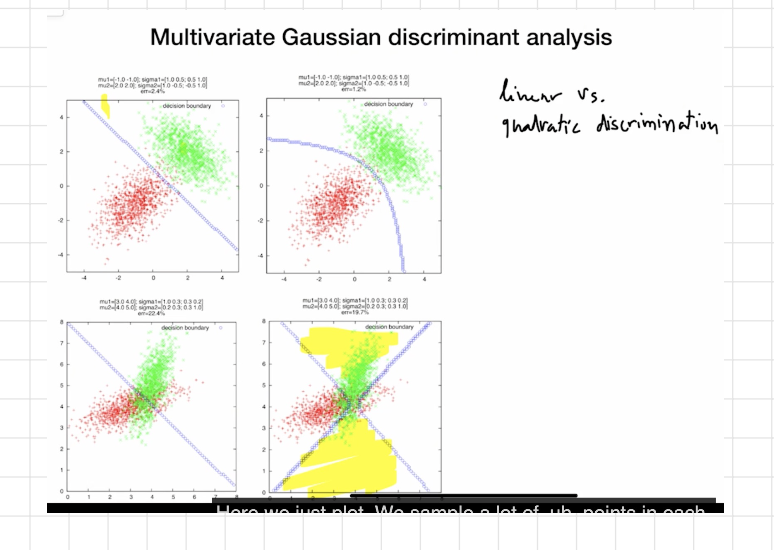
How to read your plots
Linear panel: same Σ for both classes → straight boundary.
Quadratic panel: different Σk (e.g., one class more elongated/tilted) → boundary bends; you can even get multiple disjoint regions in 1D.
TL;DR
Different covariances → QDA → Quadratic boundaries.
Same covariance → LDA → Linear boundaries.
Same covariance + priors → pick the closest mean (Euclidean if spherical).
js image

estimating distribution parameters
maximum likelihood (MLE)
What we’re estimating (per class k)
Prior πk=P(y=k) → how common the class is
Mean μk → average feature vector in class k
Covariance Σk→ spread/shape of features in class k
Pick parameters θ that make the observed data most probable:
θ^=argθmax L(θ), L(θ)=P(all data∣θ).
With the standard i.i.d. assumption (examples are independent and identically distributed):
L(θ)= i=1∏m P(x(i),y(i)∣θ).
Independence ⇒ product over i.
Identical ⇒ each term uses the same parameters θ
We almost always maximize the log-likelihood (sums are easier than products):
ℓ(θ) = logL(θ) =i=1∑m logP (x(i),y(i) | θ).
estimating distribution parameters
Goal:
Estimate the Gaussian parameters for each class k: the mean μk and variance σk² (and the prior πk by Maximum Likelihood (MLE).