L7: (TCRS) Chemotaxis and motility
1/23
Earn XP
Description and Tags
These flashcards cover the key concepts and details related to the regulation of gene expression and bacterial motility and chemotaxis as discussed in the lecture.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
24 Terms
Why is motility important for bacteria?
It contributes to virulence and colonization, enabling movement toward attractants and away from repellents
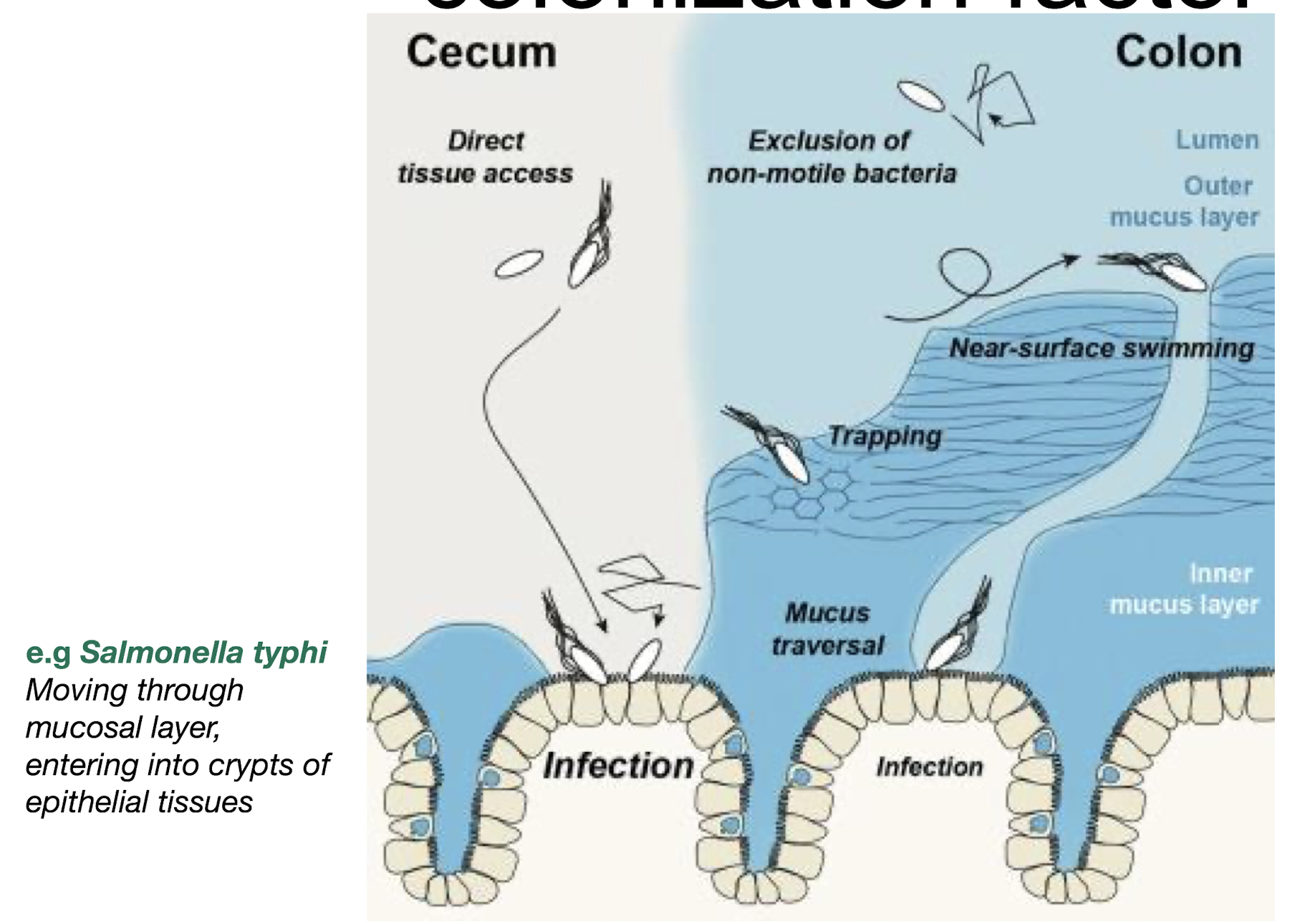
Give examples of attractants and repellents.
Attractants: sugars, amino acids.
Repellents: oxygen (for some anaerobes)
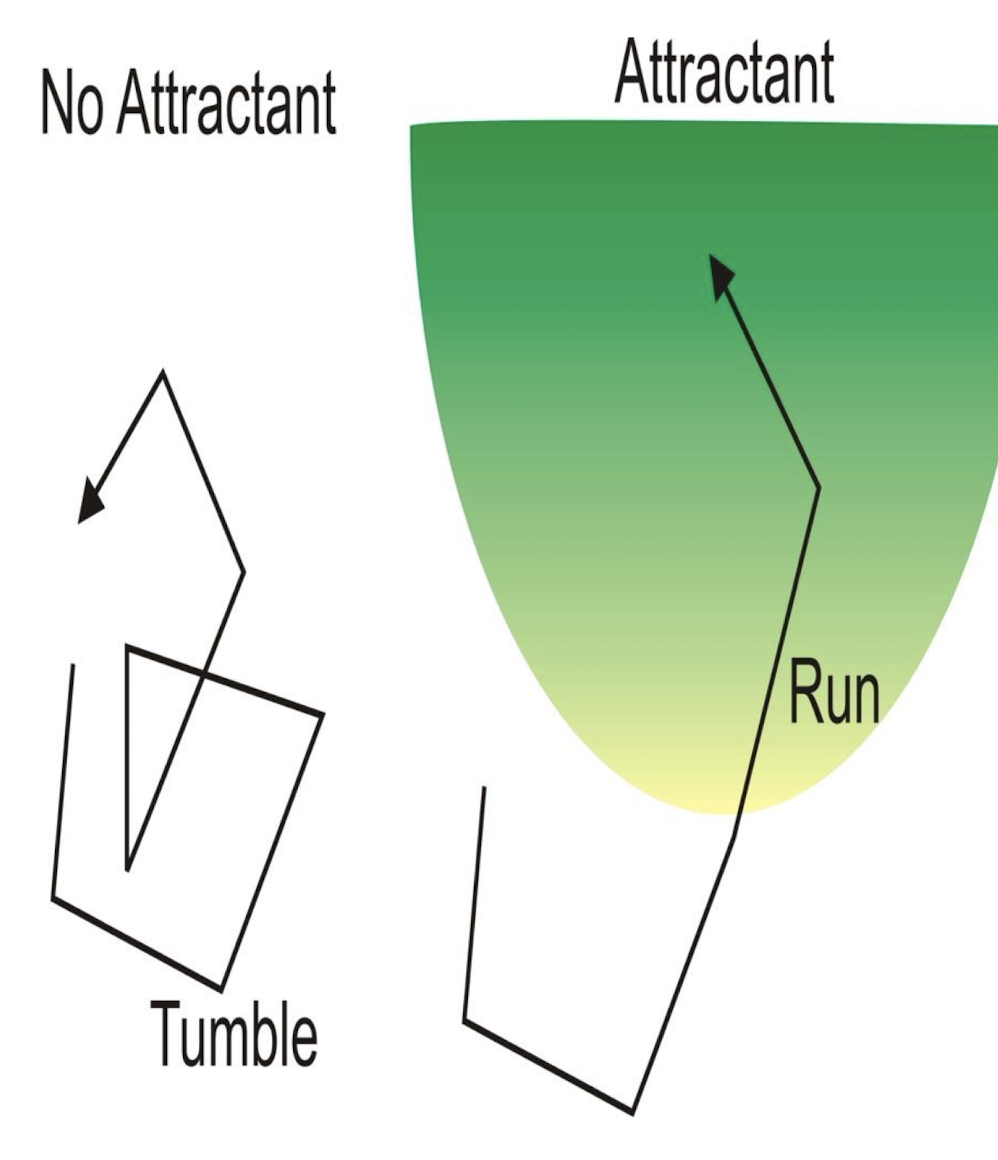
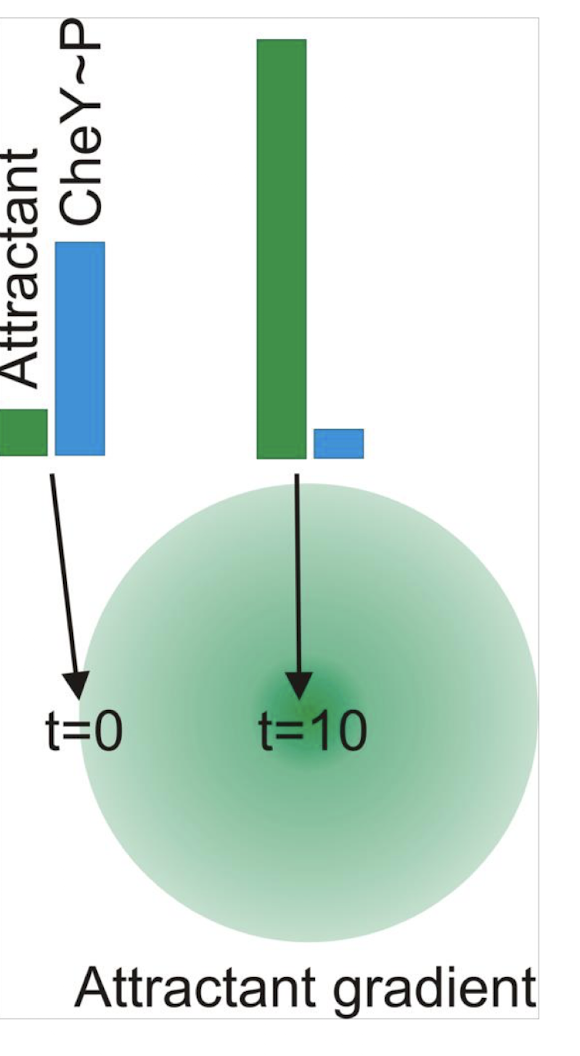
Describe random vs directed movement
Random (no attractant): frequent tumbling → random redirection
Directed (with attractant present): reduced tumbling → biased movement as long as in gradient
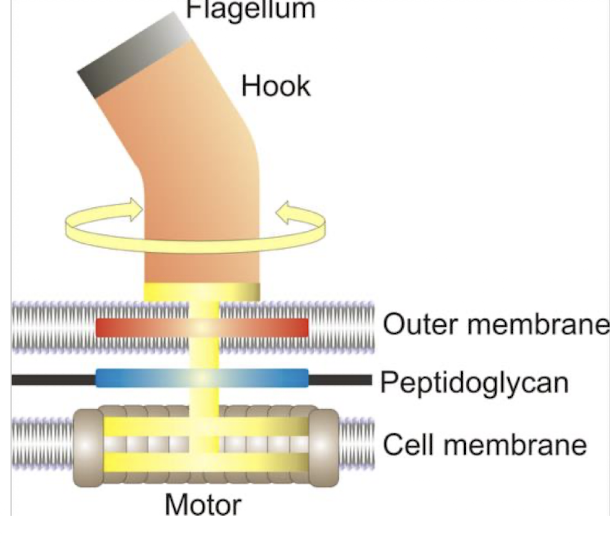
What is the flagellar hook?
A structure connecting the flagellum to the basal body, allowing for rotational movement
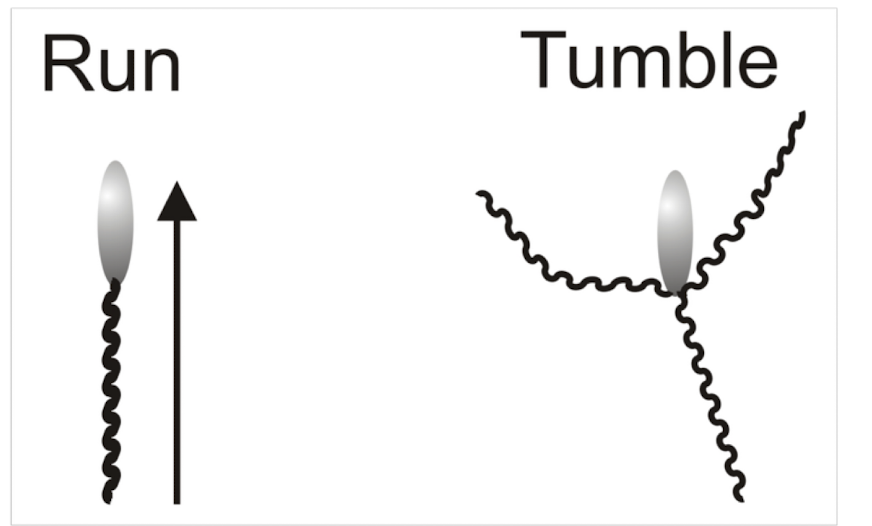
What is involved in the two gear system?
• Counter clockwise (CCW) rotation results in flagella forming bundle and bacteria moving forward smoothly
• Clockwise (CW) rotation results in flagella bundle separating and a tumbling motion being induced.
What powers bacterial flagellar rotation and how fast can bacteria move?
The rotation of bacterial flagella is powered by a proton motive force derived from the electrochemical gradient of protons across the cell membrane.
Bacteria can move at speeds up to 60 cell lengths per second.
Why do bacteria rely on temporal rather than spatial gradients?
They are too small to detect spatial difference e.g. heat in diff locations ; instead they measure changes in attractant/repellent concentration over time.
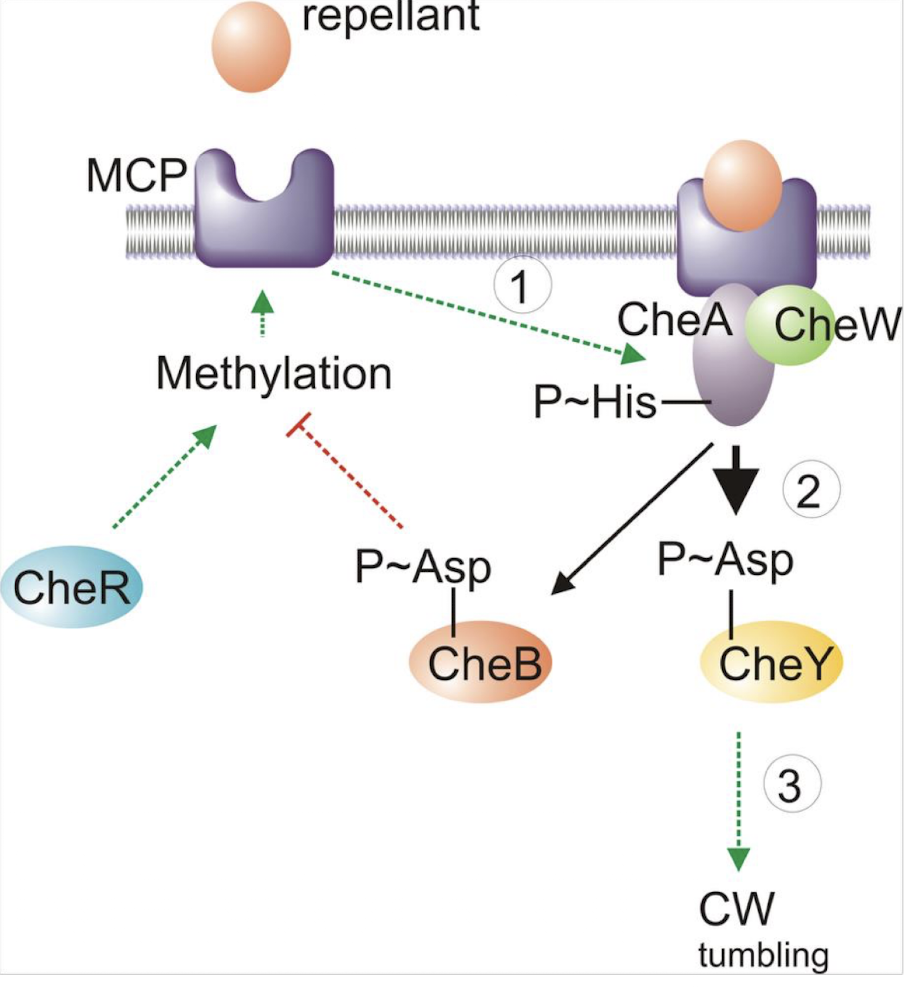
What are MCPs and what is their role?
Methyl-accepting Chemotaxis Proteins;
bind attractants/repellents
Regulate CheA autophosphorylation.
What is CheA
CheA is a histidine sensor kinase involved in bacterial chemotaxis, playing a crucial role in the phosphorylation cascade that regulates motility and sensory responses to environmental signals.
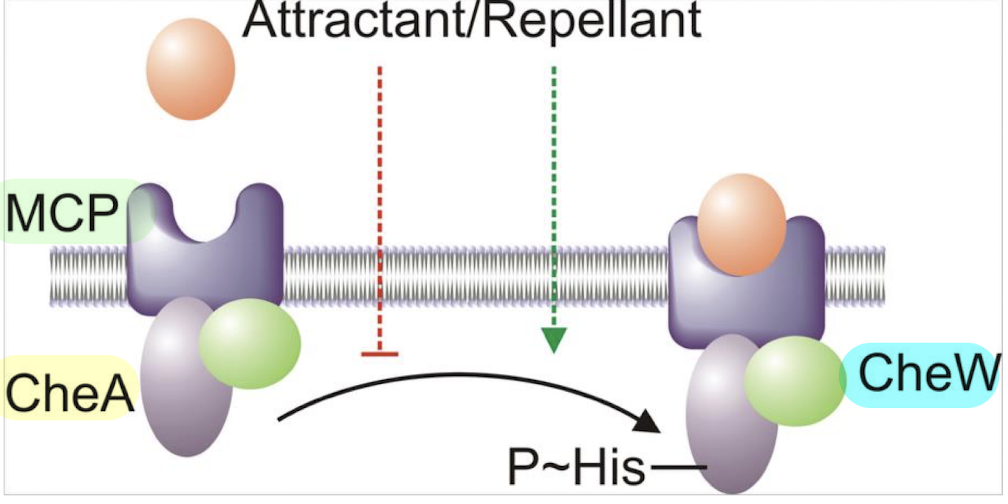
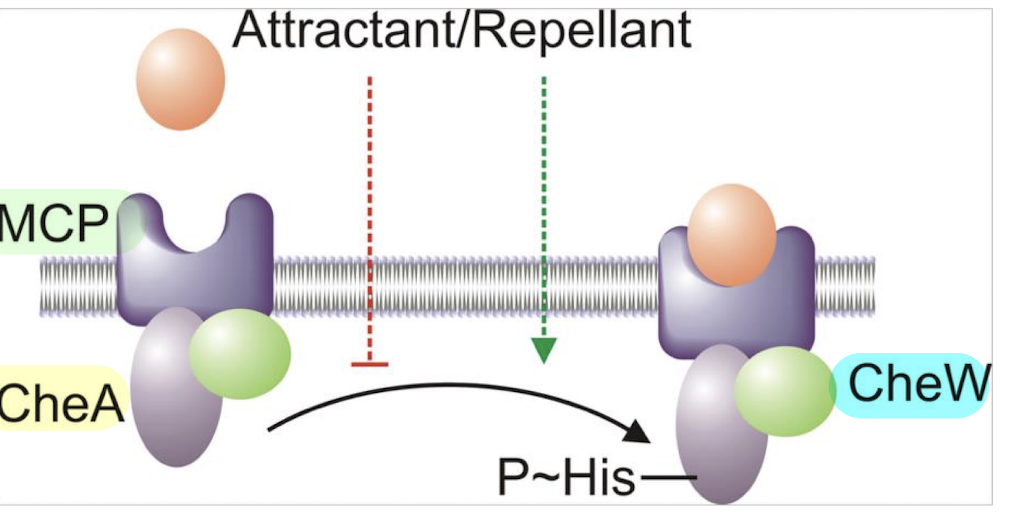
How do attractants vs repellents affect CheA autophosphorylation?
Attractants ↓ CheA autophosphorylation
Repellents ↑ CheA autophosphorylation
CheW helps couple MCPs to CheA.
What is CheW’s role?
CheW is a coupling protein (SK) that links MCPs to CheA, facilitating the transfer of signals that regulate CheA autophosphorylation in response to attractants and repellents.
What is CheY and how does it function?
A response regulator phosphorylated by SK CheA;
CheY-P interacts with the flagellar motor, inducing tumbling (CW rotation) as it has NO DNA-binding domain
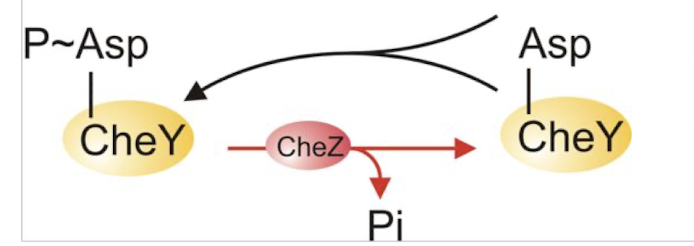
CheZ dephosphorylates CheY-P → promotes smooth swimming (CCW rotation) in bacteria.
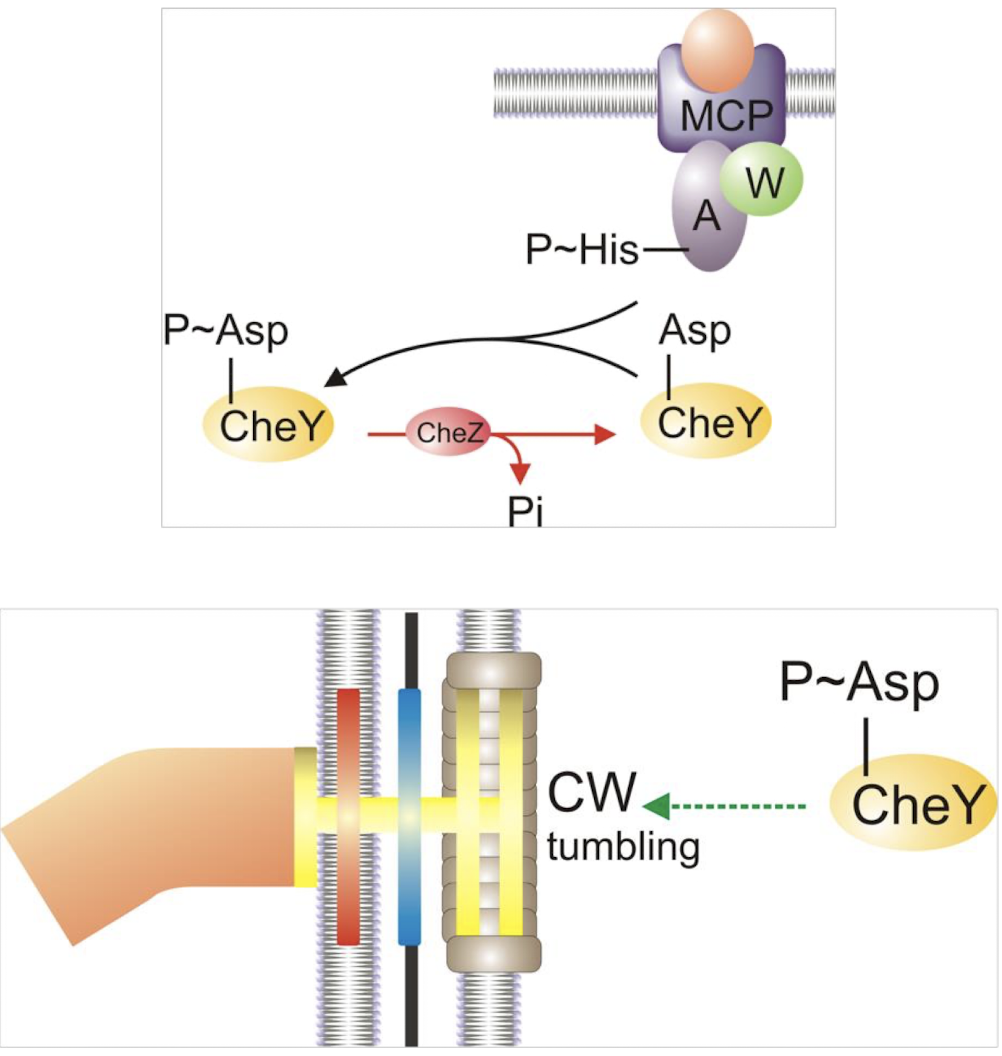
CheZ role:
can act as phosphotase to reverse process and allow to resume CCW-swimming
Repellant binding leads to _________ of CheA which _________CheY, changing flagellar rotation.
autophosphorylation which phosphorylates CheY, changing flagellar rotation - i.e. tumbling
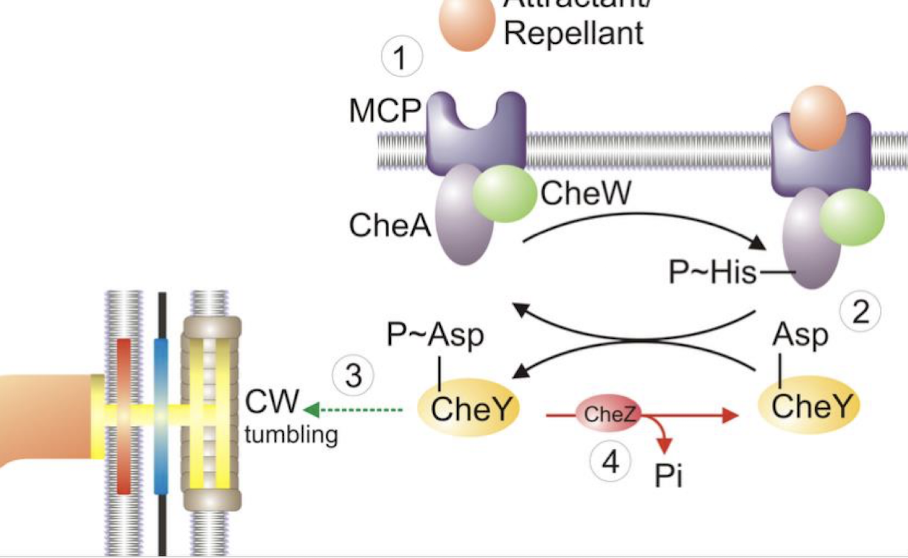
Attractant binds leading to less ___________ of CheA, which means less _________ so no change of rotation in flagella.
less autophosphorylation of CheA, phosphorylated CheY
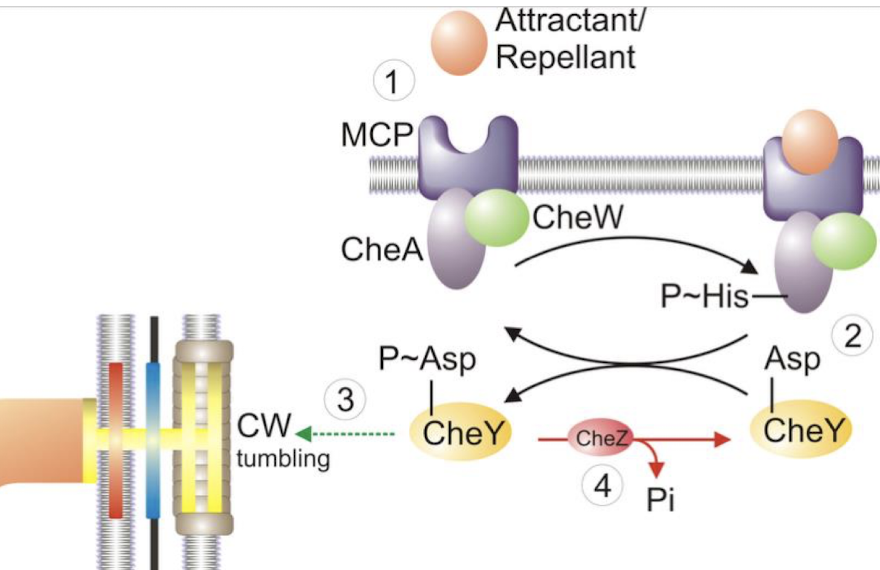
Summarize the tumbling vs swimming decision.
High CheY-P → tumbling (CW).
Low CheY-P → smooth swimming (CCW)
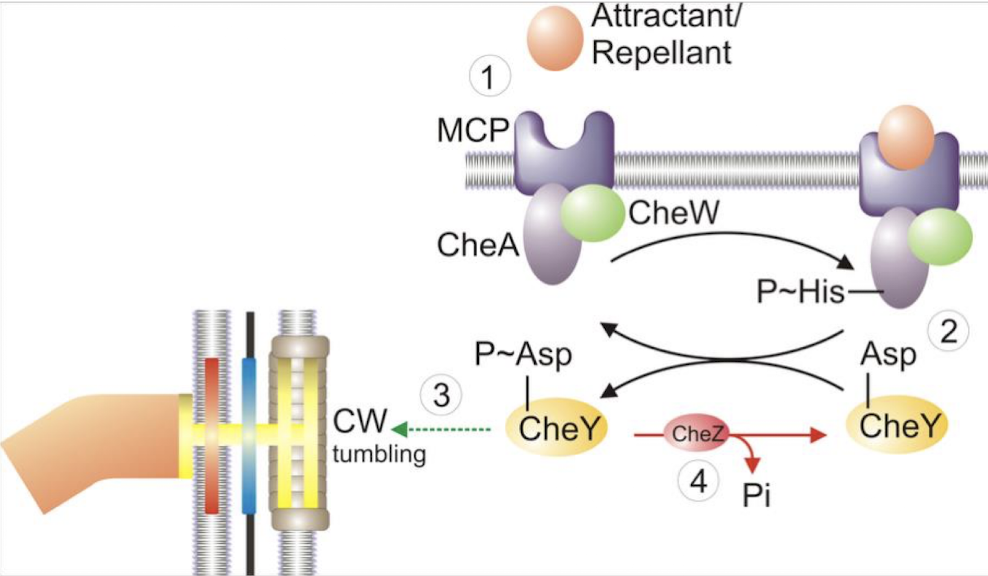
What is the flaw in the simple attractant model?
As the cell moves into the attractant, CheA-P decreases → smooth swimming continues → bacteria would “swim straight through” the attractant without stopping.
What is the flaw in the simple repellent model?
As the cell moves toward a repellent, CheA-P increases → constant tumbling → bacterium would become “stuck,” tumbling forever
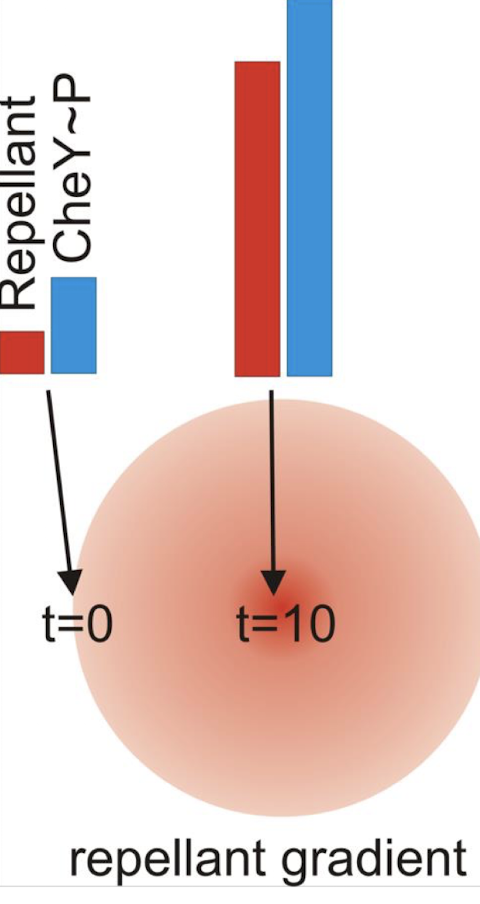
How is sensitivity in chemotaxis regulated?
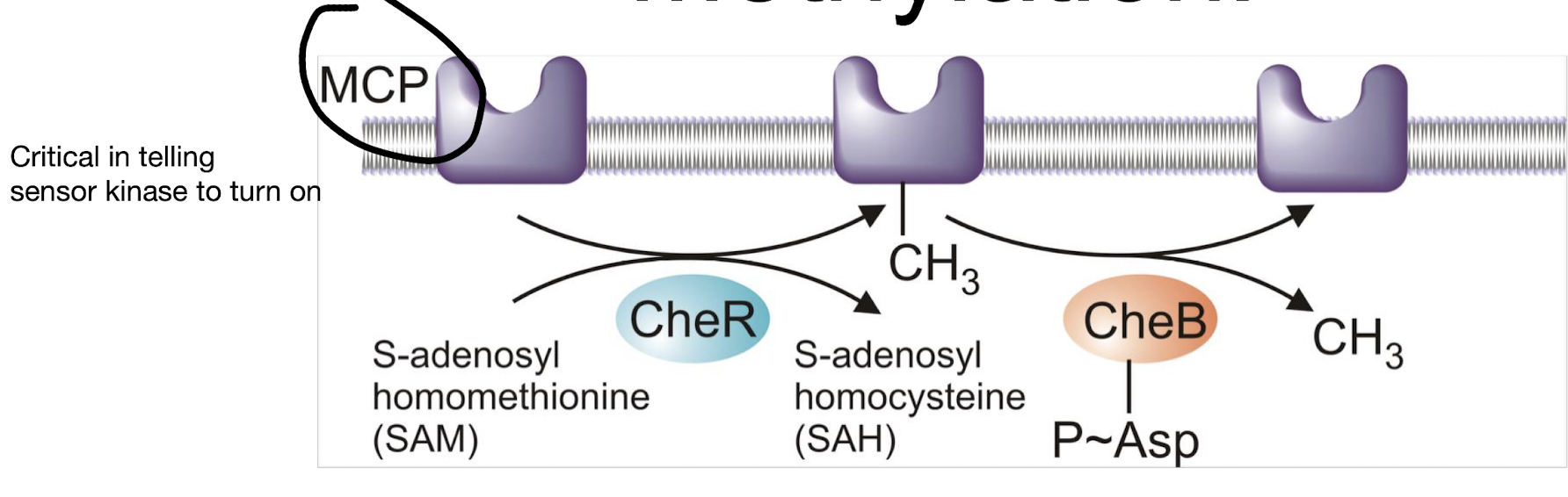
By methylation of MCPs.
CheR: a methyl transferase, slow, constant methylation, increases sensitivity.
CheB-P: a RR with methylase, decreases sensitivity
How does methylation of MCP solve the “endless tumbling” ?
Repellent binds to MCP:
CheA-P ↑ → CheY-P ↑ → tumbling.
Meanwhile, CheA slowly phosphorylates CheB-P reduces MCP sensitivity → decreases CheA signalling
→ stops excessive tumbling.
How does methylation of MCP solve the “swimming through” ?
Attractant:
Less CheB-P but high CheR methylation
System becomes more sensitive again to repellants via slower more constant methylation
prevents runaway swimming
How do TCRS regulate chemotaxis even though chemotaxis does not involve transcriptional regulation?
Although many TCRS influence transcription, the chemotaxis TCRS (CheA/CheY) modifies flagellar motor behavior, not gene expression.
Uses phosphorylation cascades to switch between smooth swimming and tumbling
Explain how motility & chemotaxis are regulated by a TCRS
The TCRS includes:
CheA (sensor kinase) → autophosphorylates depending on MCP ligand binding.
CheY (response regulator) → receives phosphate from CheA; CheY-P binds flagellar motor → clockwise rotation (tumble).
CheZ (phosphatase) reverses CheY-P and restores counter-clockwise rotation (smooth swimming).
Regulation occurs by modulating CheA phosphorylation, which directly dictates flagellar behaviour.
What are the major take-home points of the lecture?
Motility & chemotaxis rely on a two-component regulatory system without a transcriptional regulator.
MCPs, phosphorylation (CheA, CheY), and methylation (CheR, CheB-P) generate adaptive movement.
Quorum sensing allows population-dependent gene regulation via HSLs and LuxR.