chapter 3 nucleic acids
1/139
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
140 Terms
DNA and RNA are formed from
nucleotides that are linked together through a phosphodiester backbone
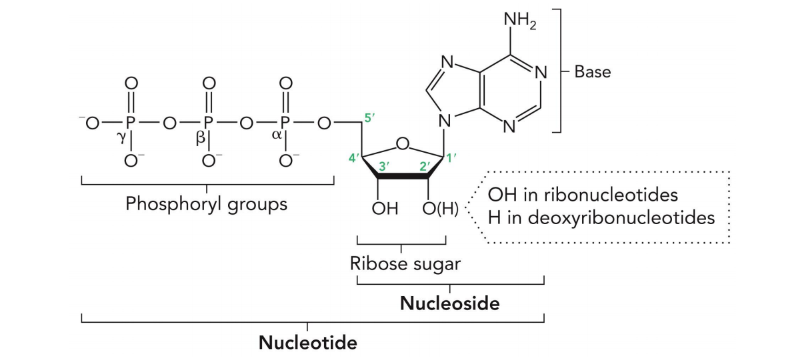
nucleotides are
phosphorylated nucleosides
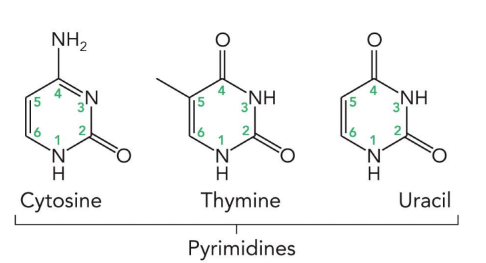
purines
adenine and guanine (9 atoms in a heterocyclic ring)
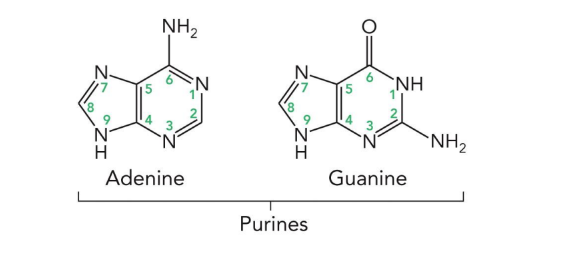
adenine
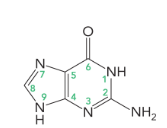
guanine
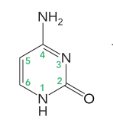
cytosine
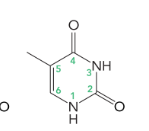
thymine
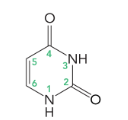
uracil
pyrimidines
cytosine, thymine, and uracil (6 atoms in a ring)
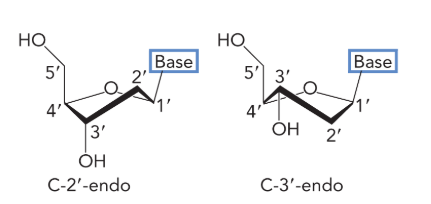
the furanose exists in one of two minimum energy conformations called
C-2’ endo and C-3’ endo
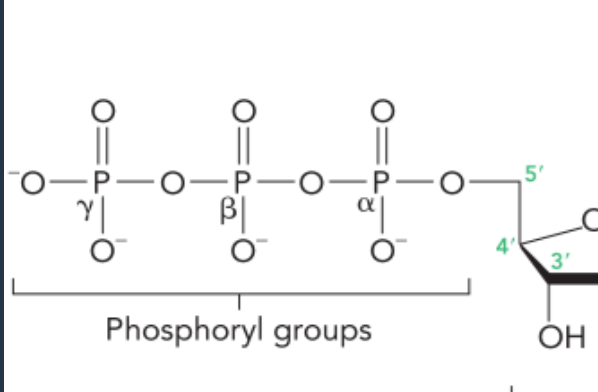
the phosphoryl groups are labelled
alpha, beta, gamma starting from the phosphoryl group closest to the sugar
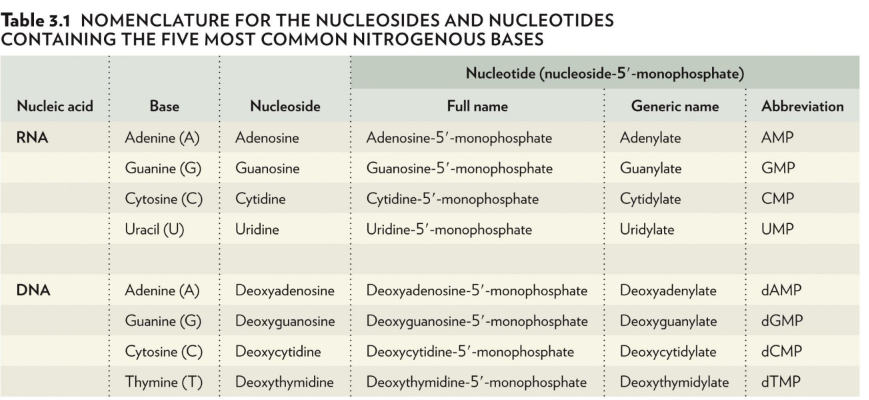
nomenclature
nitrogenous bases end in -ine
nucleosides end in -osine or -idine
full name of a nucleotide: nucleoside-5’-monophosphate (only monophosphates in DNA/RNA)
generic name ends in either -ylate or -idylate
lowercase d means it’s in DNA
the primary structure of DNA
found in all biological molecules
unique chain of deoxyribonucleotides or ribonucleotides
depicted as single letters in a row
the secondary structure of DNA
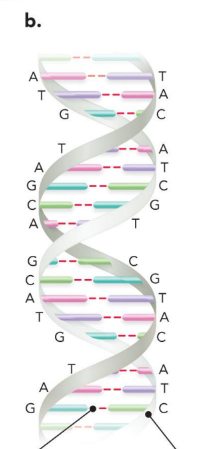
the double helix
two complementary strands of DNA annealed together in an antiparallel manner
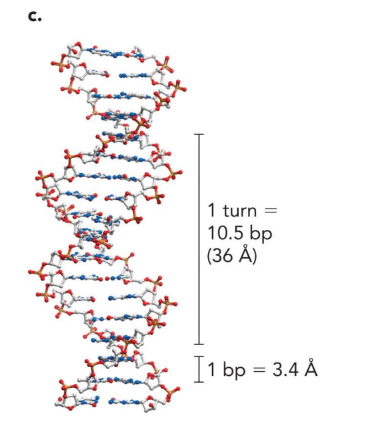
1 turn of DNA is
10.5 bp (36 angstrom)

1 bp is
3.4 angstrom
5’ has which group and 3’ has which group?
5’ phosphate
3’ hydroxyl
in ribonucleotides there is a what on 2’?
hydroxyl group
in deoxynucleotides there is a what on 2’?
a hydrogen (deoxy means no oxygen)
in order to fit within the cell, DNA must be
condensed
minor/ major groove
small distance between phosphate backbones vs large distance between phosphate backbones
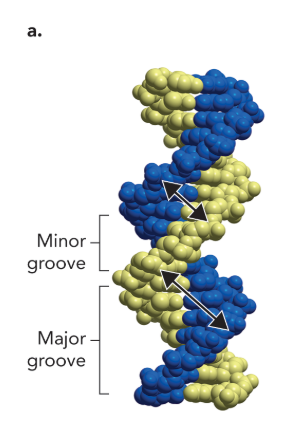
the major groove is often where
proteins specifically bind to DNA
chargaff’s rule
in the DNA from any cell of any organism, the percentage of adenine equals the percentage of thymine, and the percentage of guanine equals the percentage of cytosine
watson-crick base pairs
used the data from chargaff’s rule to propose that A paired with T and C paired with G
the base pairs are held together with
hydrogen bonds
(three form between guanine and cytosine, while two form between adenine and thymine, as well as two between adenine and uracil)
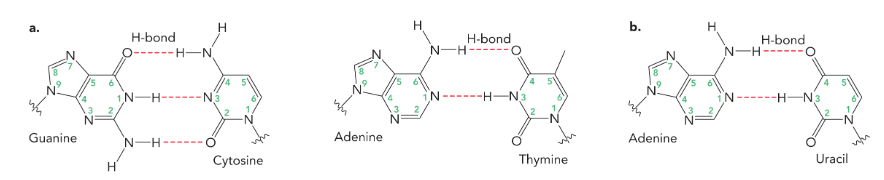
in order for a base to hydrogen bond with another base they must be
arranged in a planar fashion, parallel to the adjacent base on the same strand, and located in the interior of the helix
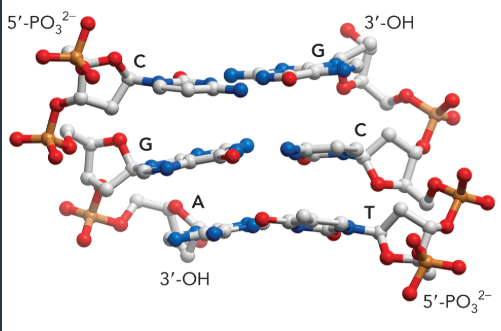
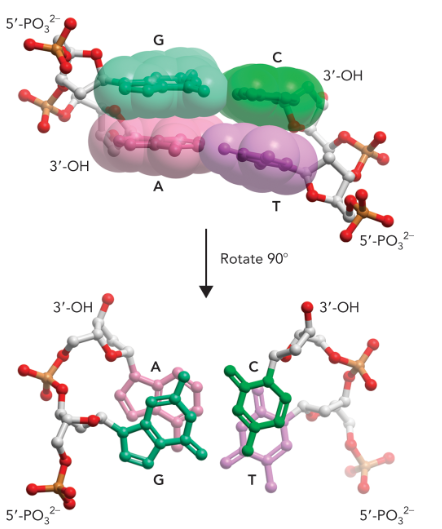
the double helix is stabilized through
base stacking at a van der waals distance which is enhanced through the hydrophobic effect (hydrogen bonding only plays a little part in stability, it’s mostly the base stacking)
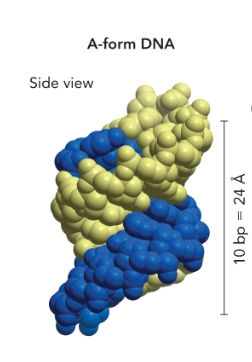
A-form DNA
short and wide
right handed
dehydrated (cannot bind to water easily)
B-form DNA
most stable
right handed

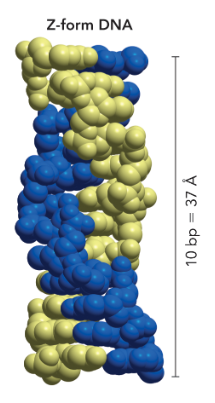
Z-form DNA
most narrow
left handed
DNA does not have a perfectly
regular or identical structure as there may be regions within a stretch of DNA more closely resembling A- or Z-DNA, depending on the sequence and presence of protein factors
it is possible for B-DNA to
transition to A- or Z-DNA without extreme changes in environment
strand separation allows for
DNA replication or transcription
in the cell, strand separation is carried out by the enzyme
helicase
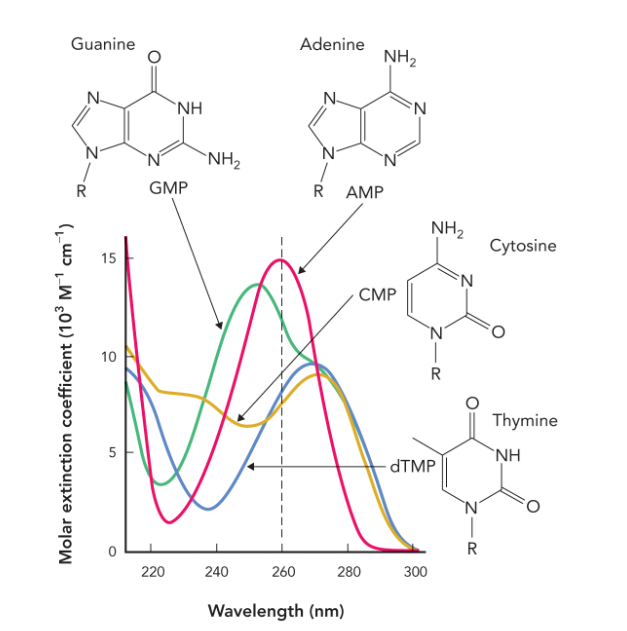
all nucleotide bases consist of
aromatic rings that absorb light in the UV range— all nucleic acids containing these bases also absorb UV light
wavelength that has a strong absorbance for mixtures of nucleotides/ DNA molecules
260 nm
hyperchromic effect
the increase in light absorbance at 260 nm as double-stranded DNA unwinds and separates
the hyperchromic effect is used to monitor
denaturation and renaturation (as DNA unwinds and denatures its absorbance increases; used to determine amount of DNA that has been denatured)
denaturation
partial or complete unfolding of the conformation of a protein or nucleic acid chain (occurs under heating or addition of acid or base)
renaturation
refolding of a denatured protein or nucleic acid chain back to its native structure and function
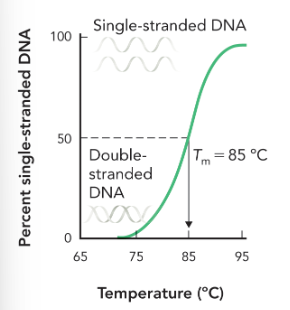
melting temperature for a region of DNA
the temperature at which half of the DNA molecules are denatured to single-stranded state, while half of the molecules are double stranded
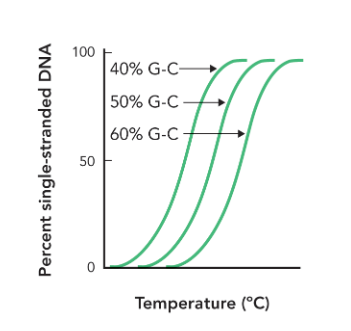
sequences with numerous A-T base pairs are more easily
disrupted and less heat energy is required to dissociate due to the less favorable base stacking interactions
as the G-C content increases
melting temperature increases (due to favorable base stacking not hydrogen bonding)
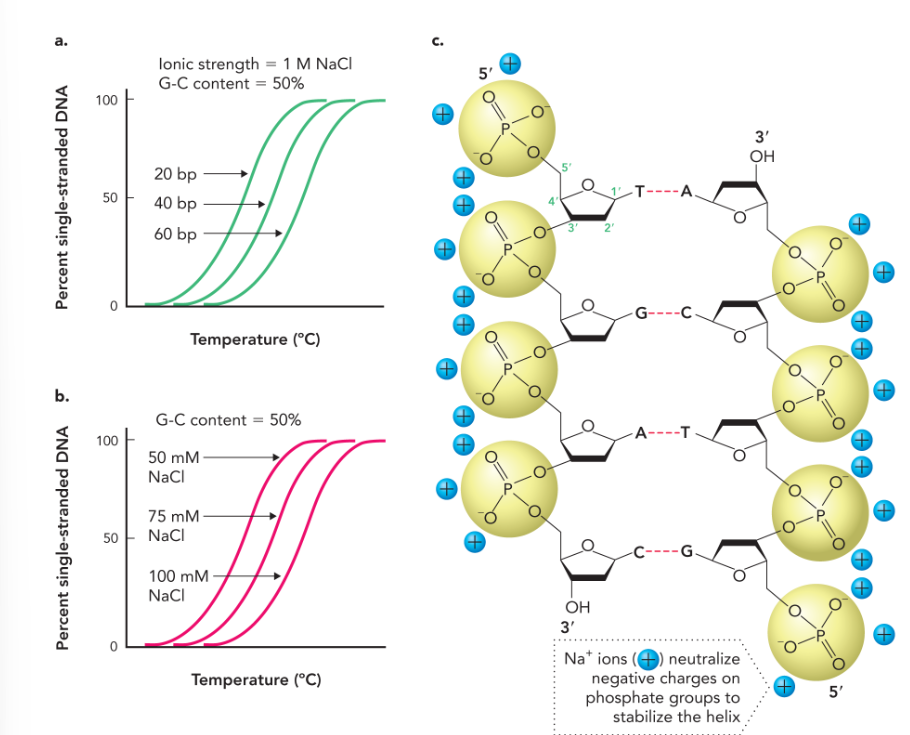
melting temperature of DNA increases with
more G-C base pairs
longer DNA strand
increased concentration of positively charged ions
supercoil
a coiled molecule, such as DNA, folded upon itself; a coiled coil
the area where the double helix crosses itself
found in prokaryotes and eukaryotes
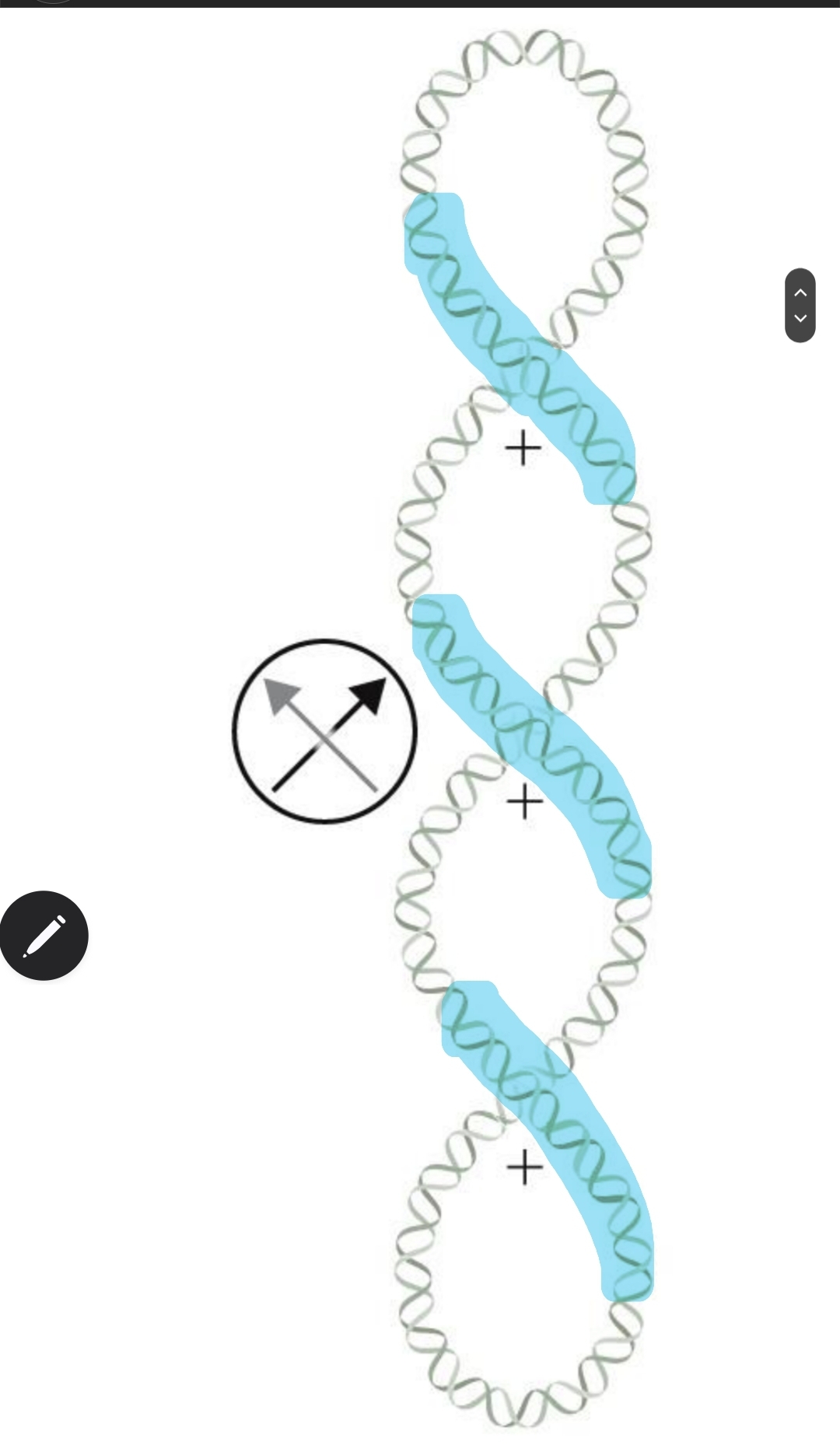
right-handed twisting results
negative supercoiling
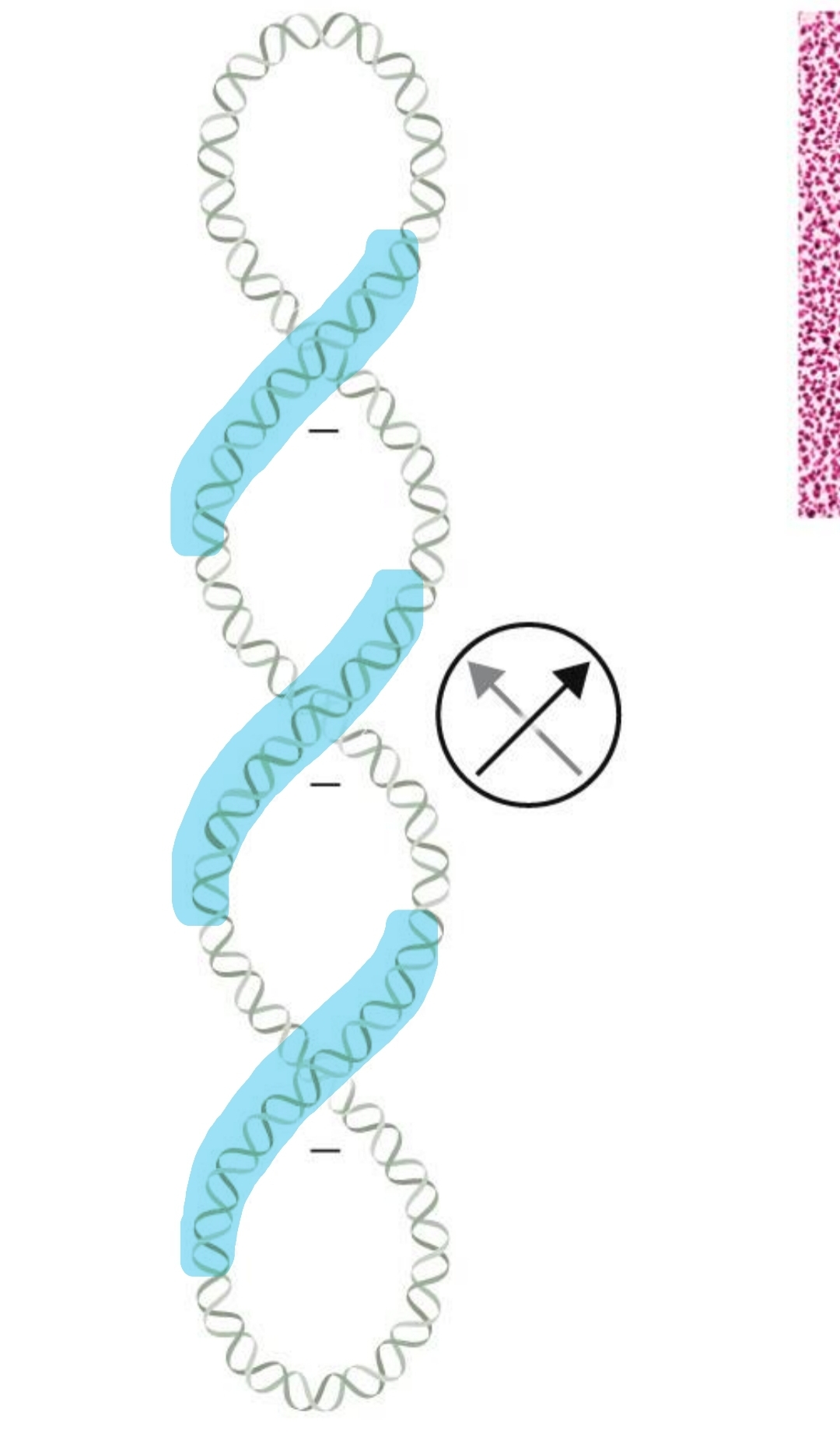
left-handed twisting results in
positive supercoiling
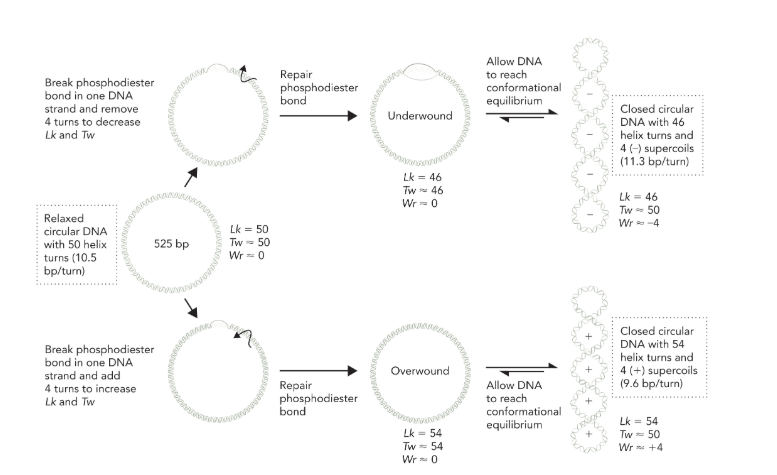
linking number
the number of times a strand of DNA winds in the right handed direction around the helix axis when the axis lies in an imaginary plane (as long as the DNA backbone is not disrupted, total Lk remains constant); Lk is calculated by dividing the total number of base pairs by 10.5
Lk = Tw + Wr
only twist and writhe change values, Lk doesn’t change unless the strand is cleaved and turns are removed or added
write changes with supercoiling
removing turns = decreasing Lk = negative supercoils = twist increases and writhe is negative
adding turns = increasing Lk = positive supercoils = twist decreases and writhe is positive
twist is just the number of turns
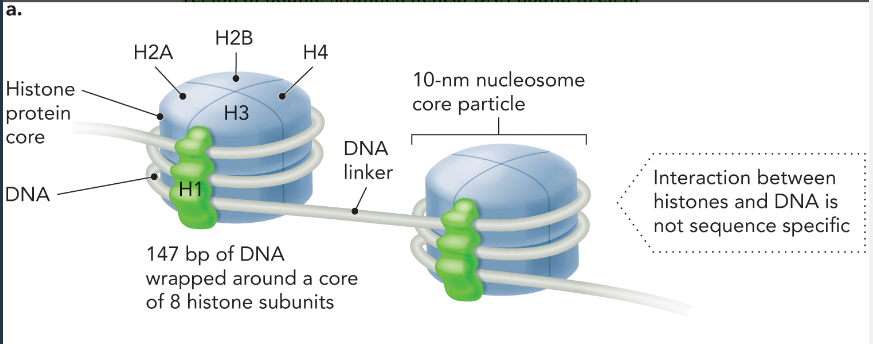
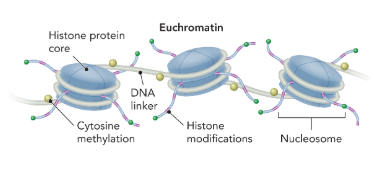
histone protein
a group of small basic eukaryotic proteins around which DNA wraps to form nucleosomes (where supercoiling occurs)
DNA-histone protein interaction
147 bp of DNA wrap around a core of 8 histone subunits to form a nucleosome (2x H2A, H2B, H4, and H1)
there is a DNA linker in between each nucleosome
each nucleosome is 10 nm
not sequence specific
circular DNA wrapping around a nucleosome induces
a negative and positive supercoil
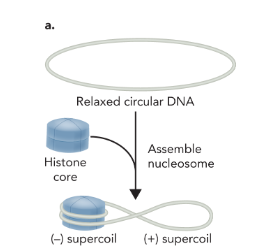
topoisomerase
an enzyme that catalyzes the cleavage of one or both DNA strands and relaxes positive supercoiled regions, allowing DNA to return to its relaxed state
to relieve the positive supercoil produced by circular DNA wrapping around the histone complex
topoisomerase cleaves and reseals the DNA which leaves the negative supercoil intact
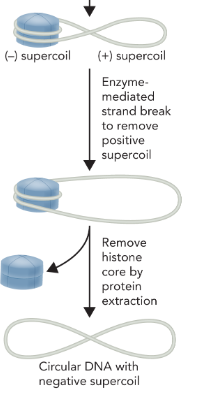
the presence of negative supercoils is advantageous to both DNA replication and transcription because
these processes involve unwinding and separating DNA strands, this is easier when the DNA has fewer turns (underwound DNA)
most genomic DNA exists in a
negatively supercoiled state
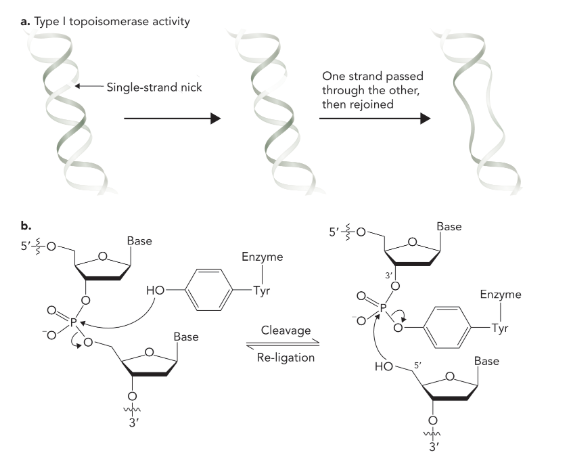
topoisomerase I
cleaves one strand of DNA
reduces supercoiled region by one turn
creates negative supercoil
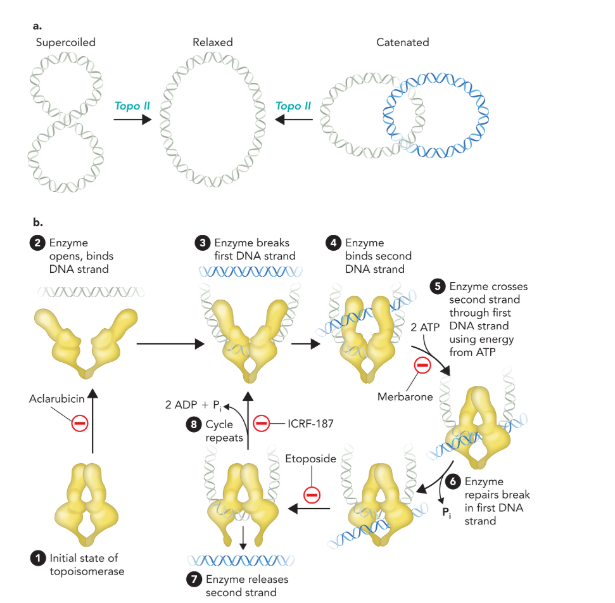
topoisomerase II
break both strands and reduces the supercoiling region by two turns; uses inhibitors to block pathways and uses energy from ATP
the presence of thymine in DNA helps to maintain
genetic information (bc of the spontaneous deamination of cytosine to uracil)
spontaneous deamination of cytosine to uracil
after the deamination adenine will be inserted on the opposite strand during the first round of DNA replication to base-pair with uracil (if uracil is not removed)
in the second round thymine replaces uracil
this coverts the original C-G base pair to a T-A base pair
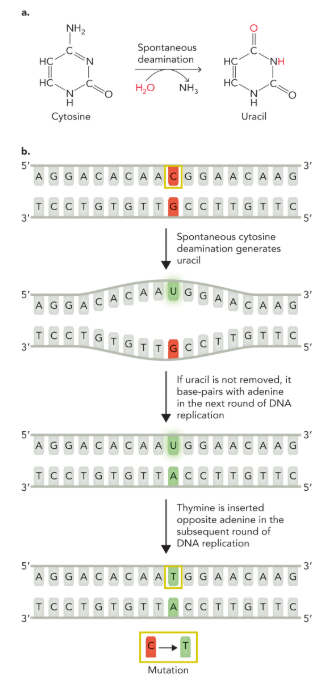
failure to remove uracil from DNA after spontaneous cytosine deamination significantly
increases the chance of accumulating deleterious mutations
ribozyme
RNA molecules with catalytic activity
ribonuclease P (RNAse P)
cleaves nucleic acids
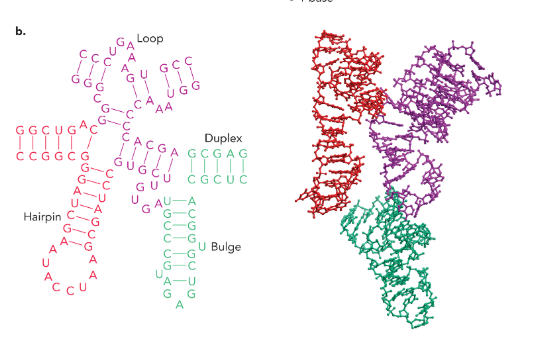
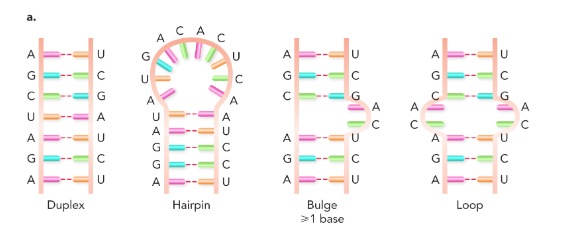
different types of RNA secondary structure
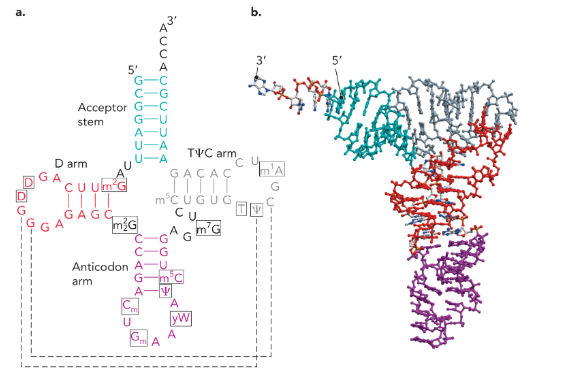
clover leaf structure of tRNA
amino-acyl tRNA
nucleosides in RNA can be
modified (guanine is the most modified)
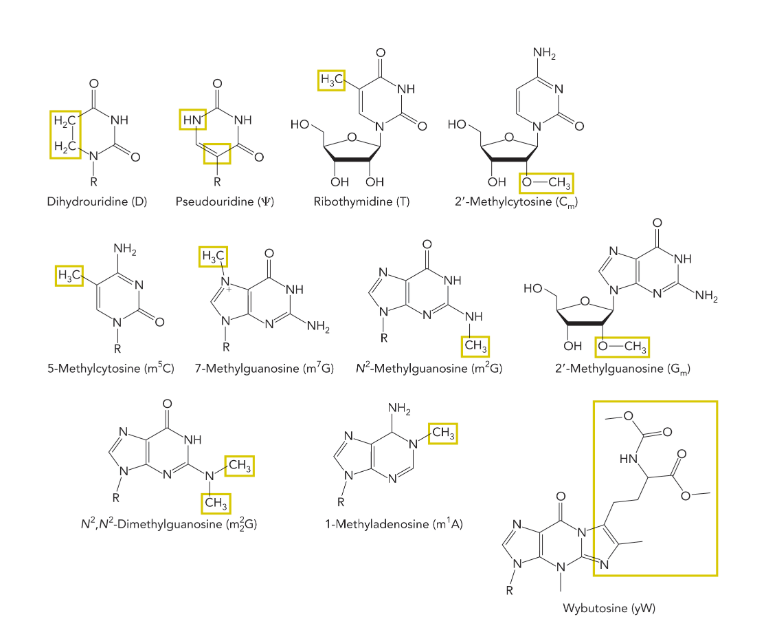
modified nucleosides in RNA molecules aid in
structure (stability) and function
many tRNA anticodons contain the modified nucleoside
inosine (able to base pair with cytidine, uridine, or adenosine; allows a single tRNA to encode the same amino acid using three different codons; means you’re most likely to have a tRNA with the right anticodon)
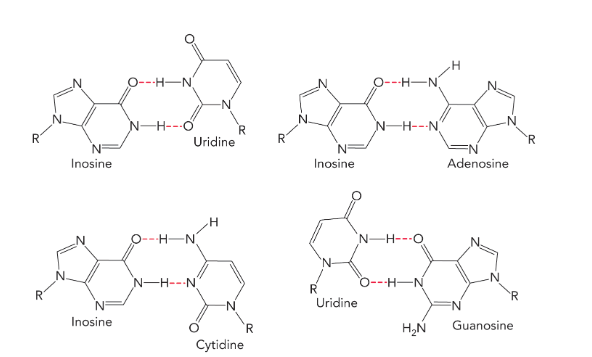
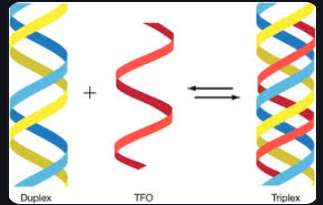
triplet interaction
occurs when a single stranded region of DNA or RNA interacts with an RNA, DNA or RNA-DNA duplex (can result in a triple helix)
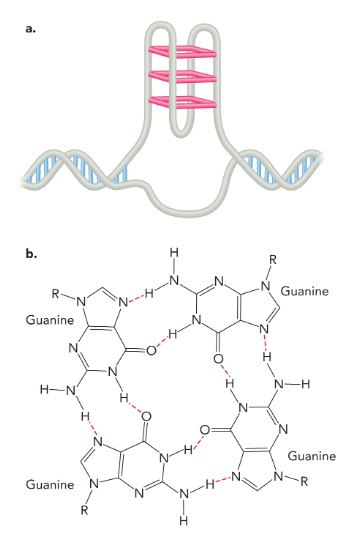
g-quadruplex
four guanine bases connected by hydrogen bonds stacked on top of each other
intercalated motif (I-motif)
hydrogen bonds between hemiprotonated cytosine residues
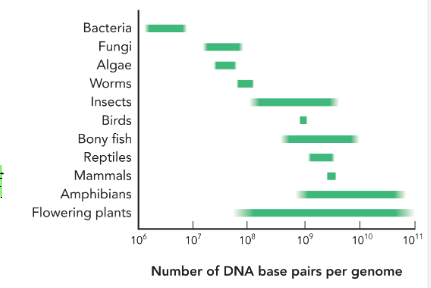
the relationship between the number of genes and the size of a genome is
not directly proportional
the number of genes increases the more
complex the organism is
genome size varies within
groups of organisms
a large amount of the human genome is transcribed into
noncoding RNA molecules
the prokaryotic genome is often contained within
a single circular chromosome that is supercoiled to fit within a cell
the number of chromosomes in an organism does not necessarily reflect
the complexity of an organism
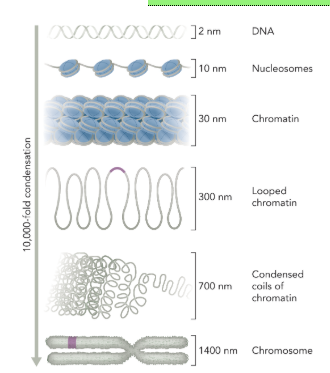
eukaryotic DNA condensation
DNA (2 nm between the strands) is wrapped around histone proteins forming a nucleosome particle (10 nm)
nucleosome particles are packed together to form a 30 nm chromatin fiber
looped chromatin (300 nm)
condensed coils of chromatin (700 nm)
chromosome (1400 nm)
from DNA to mitotic chromosome is compaction of DNA by 10,000 fold
euchromatin
a region of chromatin that is loosely packed with nucleosomes and associated with actively transcribed genes
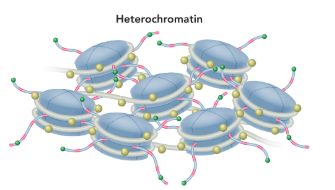
heterochromatin
a dense form of chromatin composed of mostly noncoding DNA
centromere
the region of connection between sister chromatids, composed of heterochromatin; the site of attachment for the mitotic or meiotic spindle
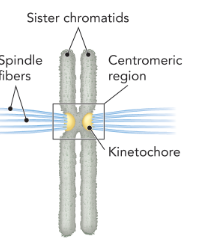
sister chromatids
two identical copies of replicated DNA contained in a mitotic chromosome
kinetochore
a protein complex, assembled at the centromere, that is necessary for proper separation of the chromosomes during cell division
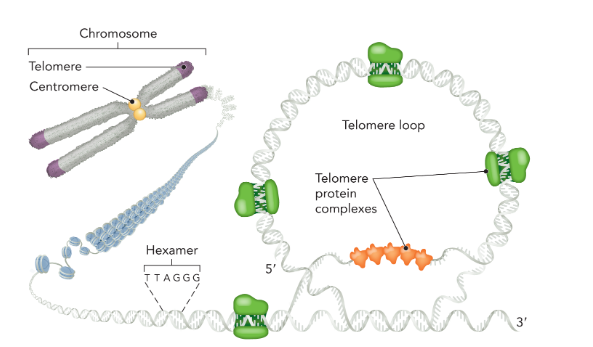
telomeres
a specialized region of heterochromatin located at the end of chromosomes; functions to maintain the length of chromosomes after replication; composed of short, repetitive DNA sequences with a high G content (5’-TTAGGG-3’) that forms loops to help protect the end of the chromosome from degradation; several protein complexes stabilize the loop structure
a gene is organized into a
promoter region and coding region
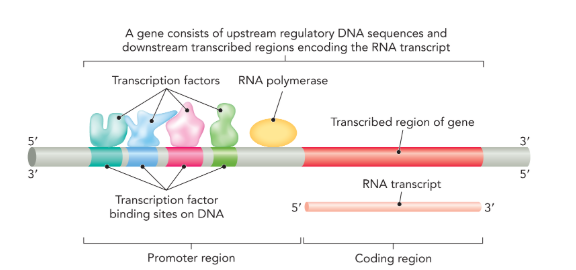
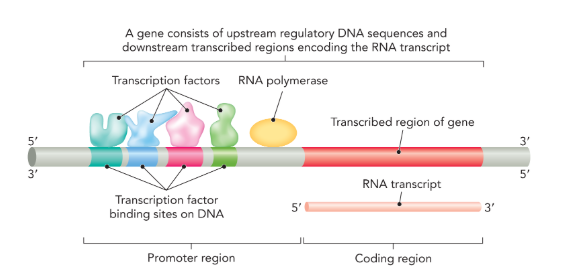
promoter region
a specific DNA sequence that occurs upstream of the coding sequence (5’); in eukaryotic cells the promoter is where transcription factors bind
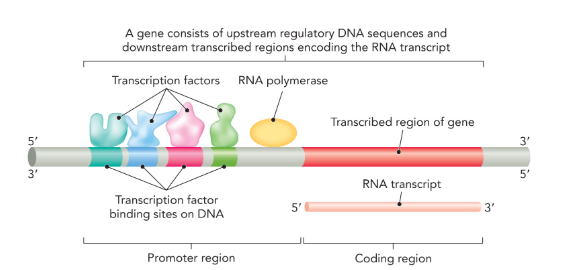
transcription factors
proteins that recognize specific DNA sequences and facilitate transcriptional initiation at gene promoters by recruiting RNA polymerase (only in eukaryotes)
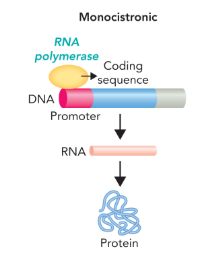
prokaryotic genes can be either
monocistronic or polycistronic
monocistronic gene
a gene that contains a promoter region followed by a single protein coding sequence
polycistronic gene
a gene that contain a promoter region followed by multiple coding regions which results in a single mRNA but multiple proteins (one from each coding regions)
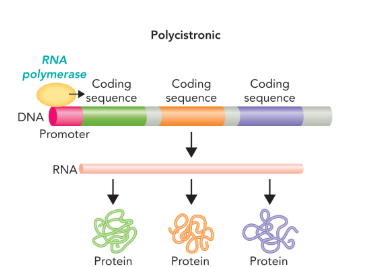
operon
a polycistronic gene that contains coding sequences for proteins involved in a single biochemical process or pathway (transcribed as a single mRNA)
in eukaryote genes coding regions called what are separated by noncoding sequences called what?
exons are separated by introns
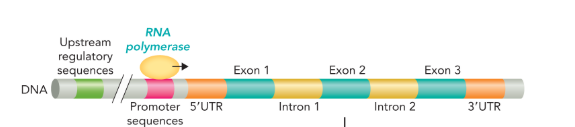
the structure of a eukaryotic gene contains the
upstream regulatory sequences, the promoter sequences, and 5’UTR upstream of the coding sequence
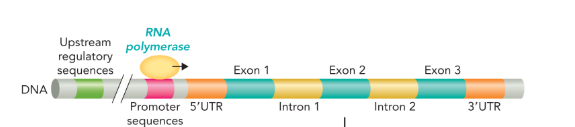
5’UTR and 3’UTR (untranslated region)
5’UTR separates the promoter region from the coding sequence
3’UTR contains sequences necessary for the termination of transcription by RNA polymerase
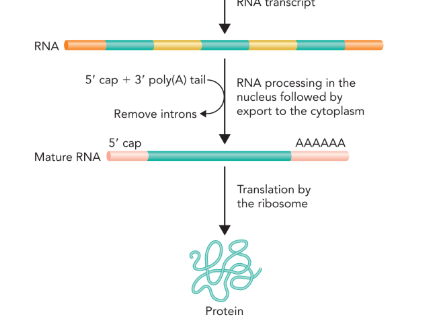
initial processing of mRNA
5’ capping— addition of methylguanosine
3’ poly-A tail (polyadenylation— addition of 100-250 adenine nucleotides)
splicing— introns are removed by a complex of proteins called spliceosome; splicing can be used by a the cell to increase the variability of a protein produced by a gene
contributes to stability and translational efficiency of mRNA
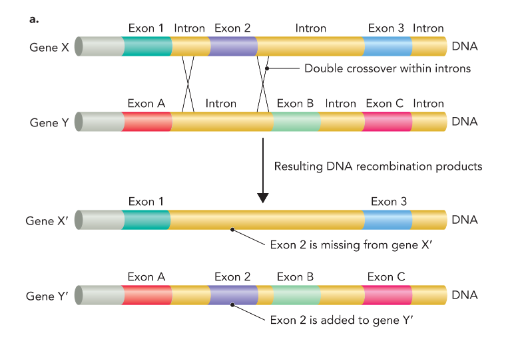
exon shuffling
the mixing and matching of protein-coding sequences during evolution to generate genes with new function
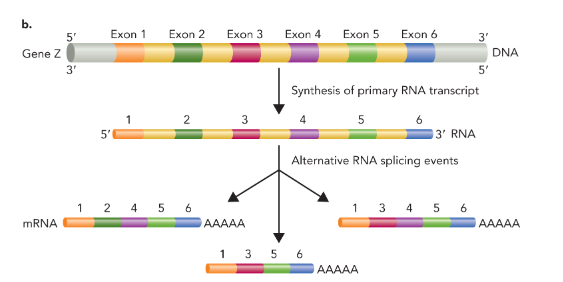
alternative splicing
combing exons in different ways to produce proteins with alternative structures and/or functions (makes multiple distinct mRNA products)
bioinformatics
uses computational tools to analyze data generated from molecular and biological samples; can be used to discover the function of an unknown gene such as human disease genes, which could then lead to drug development to target the protein
computational methods in genomics allows scientists to
find differences caused by mutation or normal genetic variation or determine how the nucleotide sequence of a gene or genomic region has changed with evolution
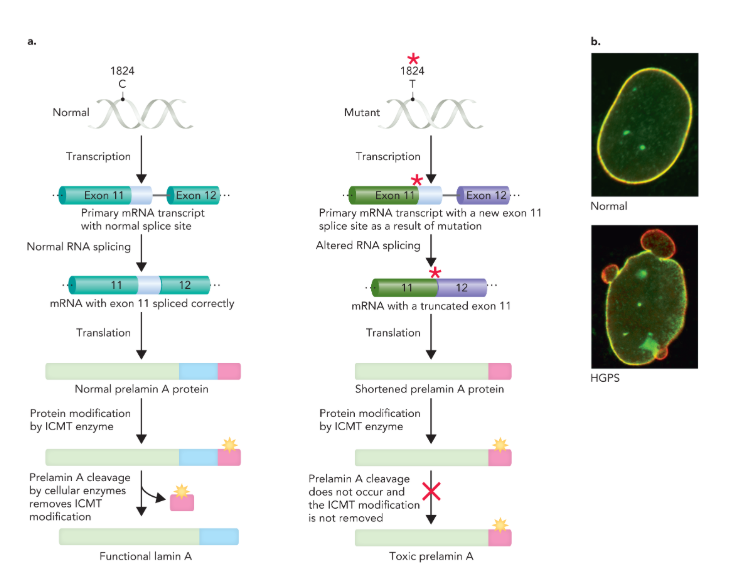
Hutchinson-Gilford progeria syndrome
fatal disorder that causes accelerated aging in children
commonly caused by a mutation in humans
changes the nucleotide at position 1824 in the coding sequence of exon 11 from C to T in the lamin A gene, which leads to a different splicing of exon 11 to exon 12
splicing leaves about 150 nucleotides missing and results in a shortened prelamin A precursor protein
precursor protein gets modified by the enzyme ICMT, which normally gets removed to create a functional lamin A protein, but in the mutant form the modification is retained and leads to the accumulation of the mutant protein in the nuclear membrane
the buildup of the altered protein damages the structure and function of nuclei, which leads to premature cell death
bioinformatics made it possible to diagnose the disease earlier and develop new potential treatments