genetics exam 1
1/228
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
229 Terms
heredity
a transmission of properties from one generation to the next
genotype
heritable information that an individual possesses
phenotype
physical, observable traits that an individual possesses
central dogma
a concept of information flow in living cells
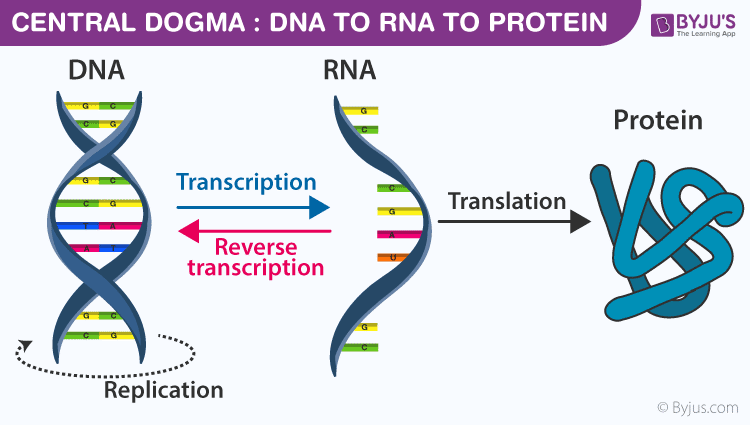
DNA
deoxyribonucleicacid; double-stranded, antiparallel molecule that carries genetic information for the development and functioning of an organism
RNA
ribonucleic acid; single-stranded; principle role is to act as a messenger between DNA and the synthesis of proteins; carries genetic information in some viruses
protein
a large molecule made up of amino acids whose sequence is determined by the DNA code within a gene
replication
the process of duplicating a strand of DNA
transcription
the process of going from DNA to RNA
translation
the process of going from RNA to proteins
prokaryote
a single-celled organism whose cells lack a nucleus and other organelles
eukaryote
an organism whose cells contain a nucleus within a membrane
transforming principle
a physical material that is transmissible and heritable
“Smooth” strain of S. pneumoniae
appears shiny and smooth due to a polysaccharide capsule that protects it from the host's immune system which makes it virulent.
“Rough” strain of S. pneumoniae
appears rough and lacks the polysaccharide capsule, making it non-virulent.
virulent organisms
those that are able to cause disease
non-virulent organisms
those that are not able to cause disease
bacteriophage
A virus that injects material inside of its host cell and uses the host cell’s components to many copies of itself. The host cell eventually lyses and the virus copies are released.
E. coli
an bacteria found in the lower intestine of warm-blooded organisms; used in the Hershey-Chase experiment
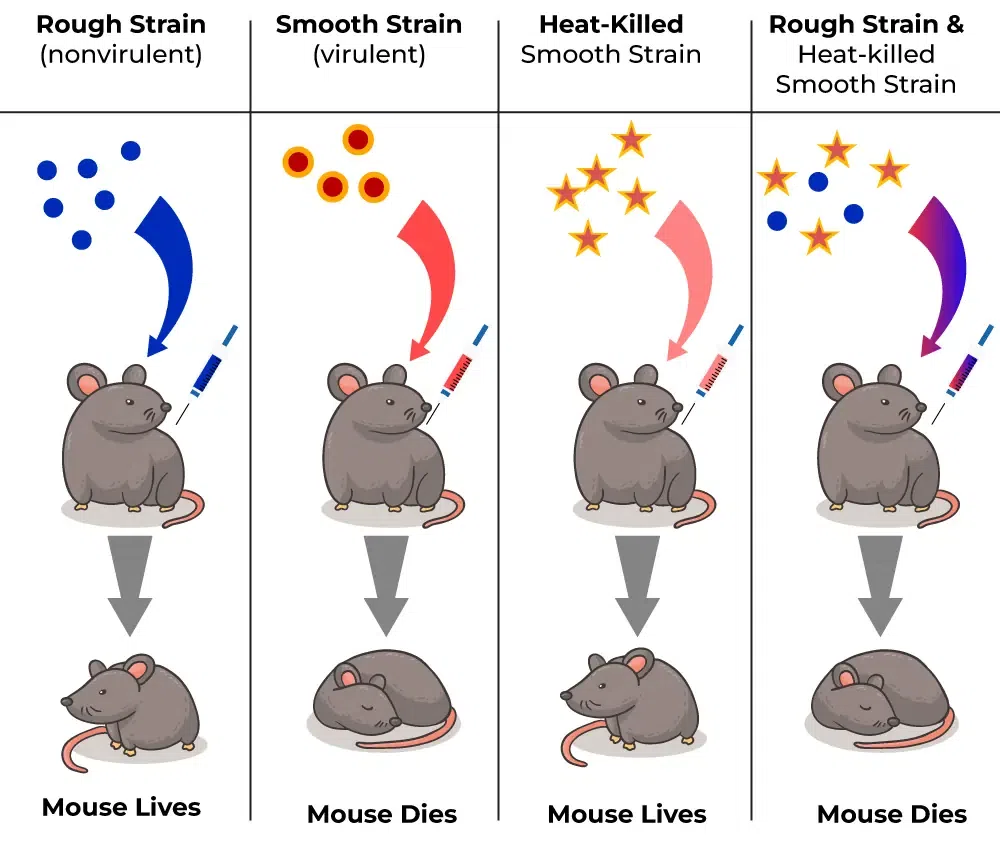
Griffith’s experiment and conclusions
The transforming principle: When a mouse was injected with a live smooth virulent of of Streptococcus pneumoniae, it developed pneumonia. When injected with heat-killed smooth strains and live rough strains, the mouse survived, but when both were combined, the mouse died, indicating transformation. This meant that the observable traits of Streptococcus pneumoniae are caused by a physical material that’s transmissible and heritable
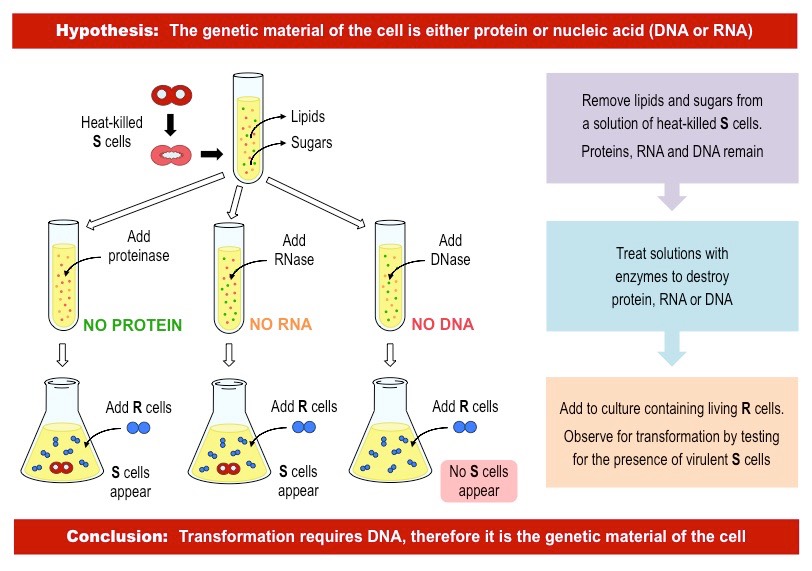
Avery, McCarty, and McLoed’s experiment and conclusions
Demonstrated that DNA is the transforming principle. They isolated DNA from heat-killed smooth strains and showed that it could transform rough strains into smooth, confirming that DNA carries genetic information. Under the same process, proteins and RNA were not capable of the transformation from smooth to rough strains.
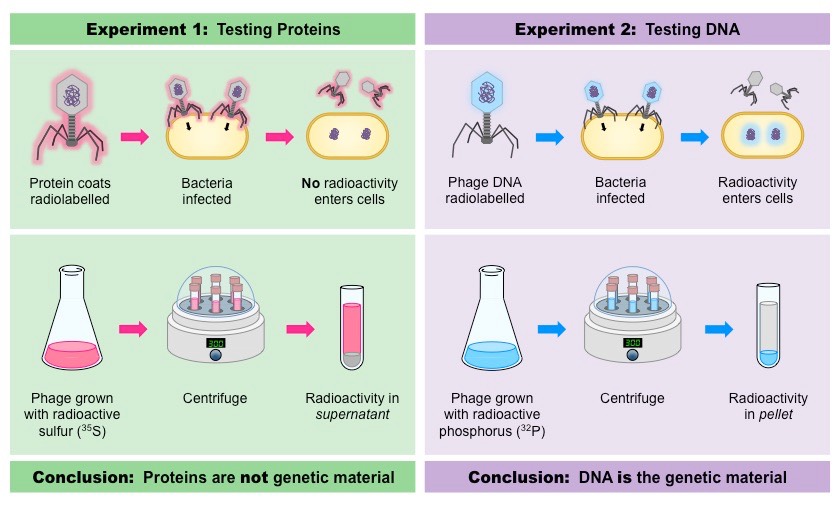
Hershey and Chase’s experiment and conclusions
Demonstrated that DNA, not protein, is the genetic material by using radioactive isotopes to label the protein and DNA components of a bacteriophage (virus that infects bacteria), then showing that only the labeled DNA entered the bacterial cell during infection.
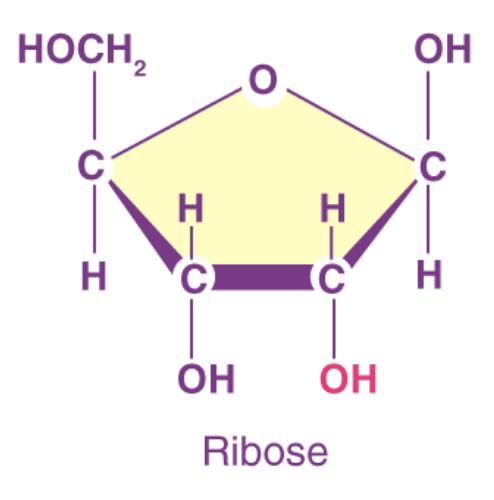
ribose
pentose sugar (OH chemical group); RNA
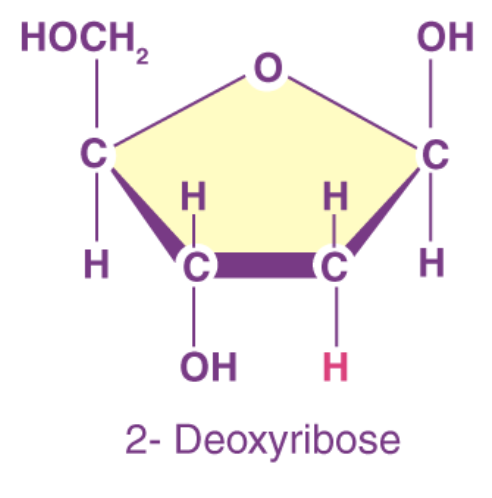
2’-deoxyribose
pentose sugar (H chemical group); DNA
nitrogenous base
covalently attached to the 1’ carbon; include purines and pyrimidines
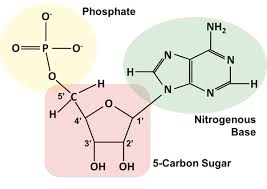
triphosphate
phosphate groups attached at the 5’ carbon; make attachments to other nucleotides via their 3’ carbon
hydrogen bonds
procures interaction between antiparallel strands through nitrogenous bases; relatively weak
What kinds of bonds are phosphodiester and hly
include phosphodiester and glycosidic bonds
phosphodiester bonds
connect phosphate group to 3’ carbon in next nucleotide to form the phosphate backbone
glycosidic bonds
connect nitrogenous base to 1’ carbon
purine
2-ring structure: 1 6-member ring, 1 5-member ring; adenine and guanine
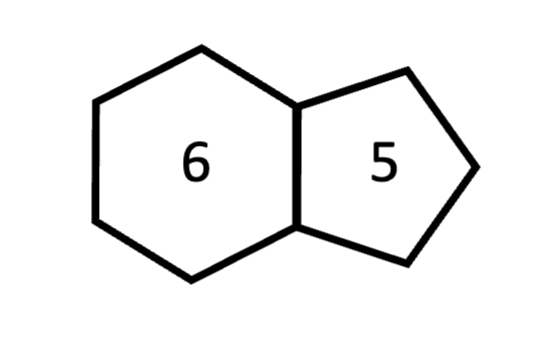
pyrimidine
1-ring structure: always 6-member ring; thymine and cytosine
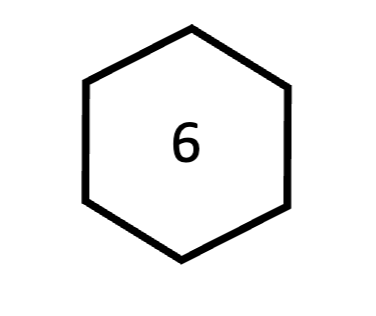
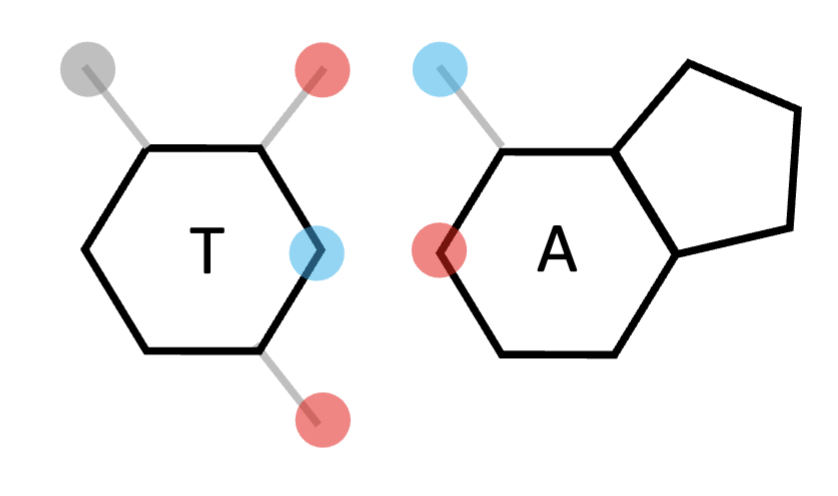
adenine
purine; forms 2 bonds
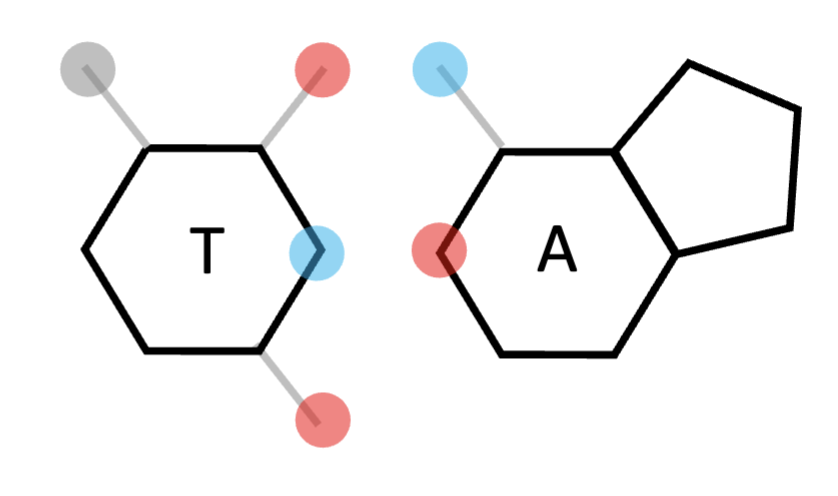
guanine
purine; forms 3 bonds
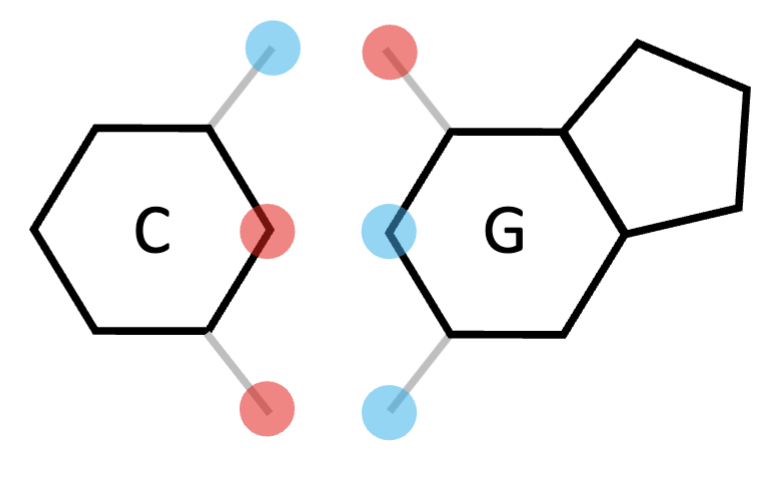
cytosine
pyrimidine; forms 3 bonds
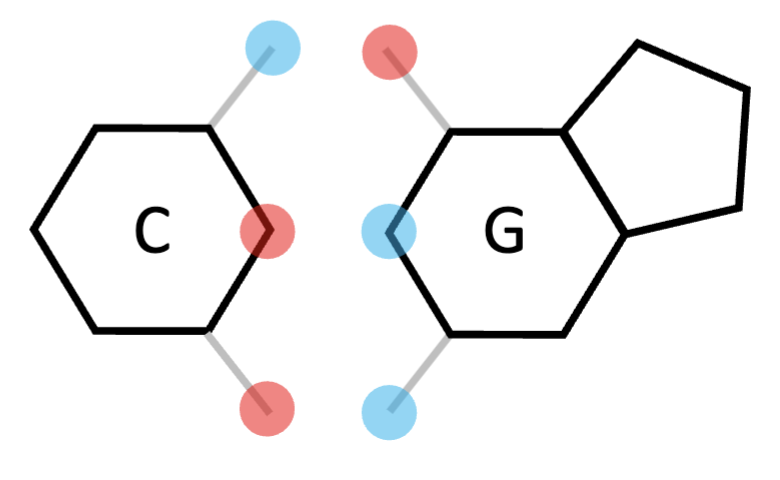
thymine
pyrimidine; forms 2 bonds
sugar-phosphate backbone
phosphodiester bonds that link nucleotides together (3’ carbon and phosphate group at 5’ carbon")
A-DNA
right-handed structure seen in conditions of low humidity; short+wide with narrow, deep major group and a wide, shallow minor groove
B-DNA
nearly all natural DNA is right-handed B-DNA; wide major groove, narrow minor groove of similar depths
Z-DNA
left-handed with alternating purine and pyrimidine bases; major groove near surface of the helix
minor groove
some proteins bind here if they are small enough
major groove
some proteins (mostly large) bind here and can interact with nitrogenous bases
Chargaff’s rules
%A=%T
%G=%C
helix properties - 1 “rung”
involves 1 nucleotide’s base pair; 0.34 nm apart
helix properties - 1 complete turn
distance of 10 rungs (10 nucleotides); includes 1 major and 1 minor groove; length of 3.4 nm; strands are antiparallel
complementary
describes the bases on two strands; why one strand contains all info needed to produce a second strand
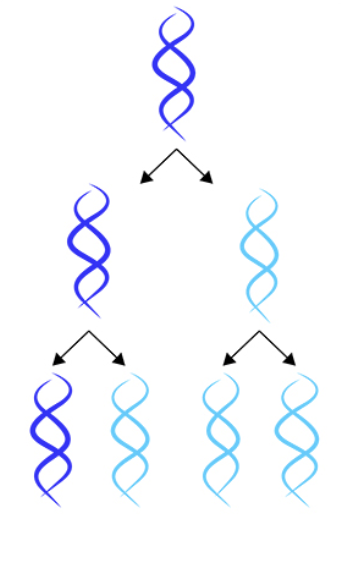
semiconservative
template hypothesis; DNA is copied to create two new DNA strands, each containing one old strand and one new strand
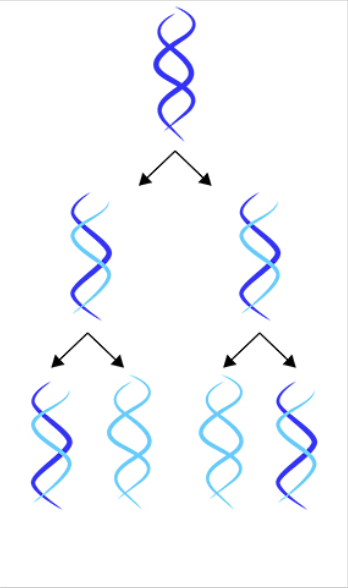
conservative
one copy is made of the old strands and the other is made of 2 new strands (conserves original strand)
dispersive
DNA replication model by which a new strand is made up of fragments from the old strand as well as newly synthesized fragments
DNA polymerase I
removes primer; also capable of DNA synthesis to replace primers
also has 3’ to 5’ exonuclease activity that enables reparations of mistakes
DNA polymerase III
responsible for most DNA synthesis; works from 5’ to 3’ starting at primer
also has 3’ to 5’ exonuclease activity that enables reparations of mistakes
template strand
single-strand DNA
primer
short, single-stranded RNA that acts as a starting point for DNA synthesis
initiator protein
opens of (melts) DNA as a step to creating a template strand
origin of replication
the location where initiator proteins perform; typically A-T rich as they are weaker (only 2 H-bonds)
helicase
responsible for unwinding DNA to create the template; work in opposite directions of the origin of replication
primase
responsible for synthesizing a short RNA primer (antiparallel to template)
ligase
connects fragments by ligating DNA at “nicks” of phosphodiester bonds from removal of primers
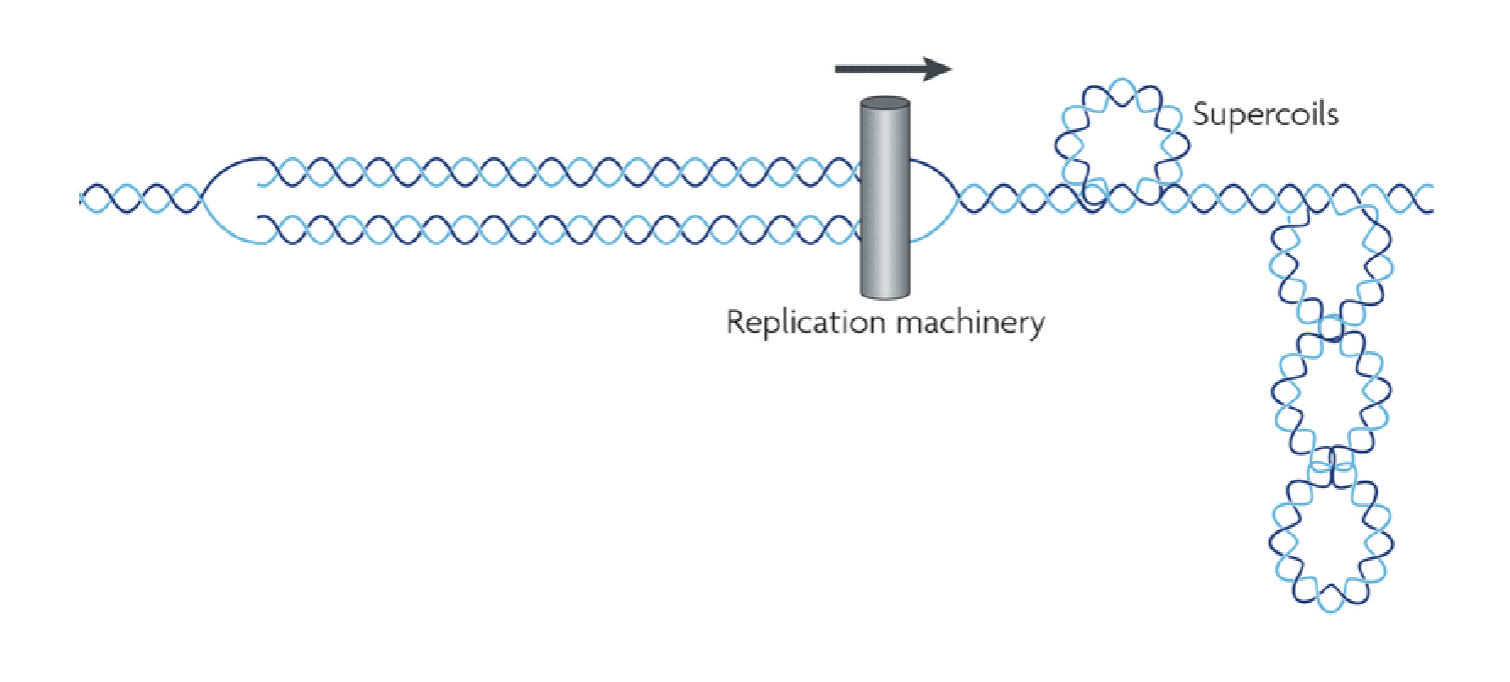
topoisomerase
relaxes supercoils caused by replication fork
telomerase
extends 3’ end to preserve the ends of a chromosome
removes RNA primer
adds an extension to the template strand
new RNA primer produced
extra DNA added
3’ to 5’ exonuclease activity
locate mismatch
remove 3’ nucleotide
add new OH, then new nucleotide
performed by DNA polymerase I and III
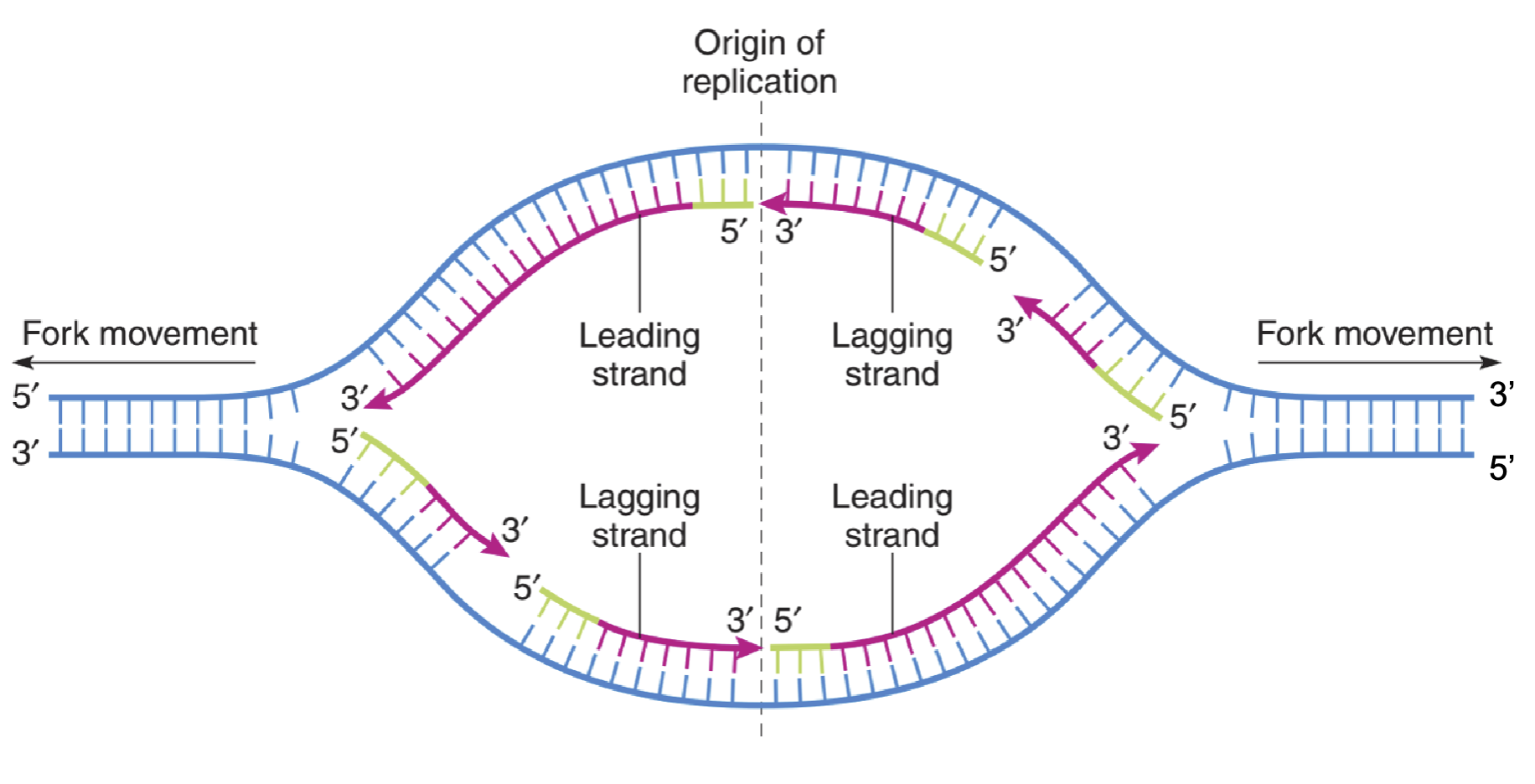
replication fork
a very active area where DNA replication processes take place
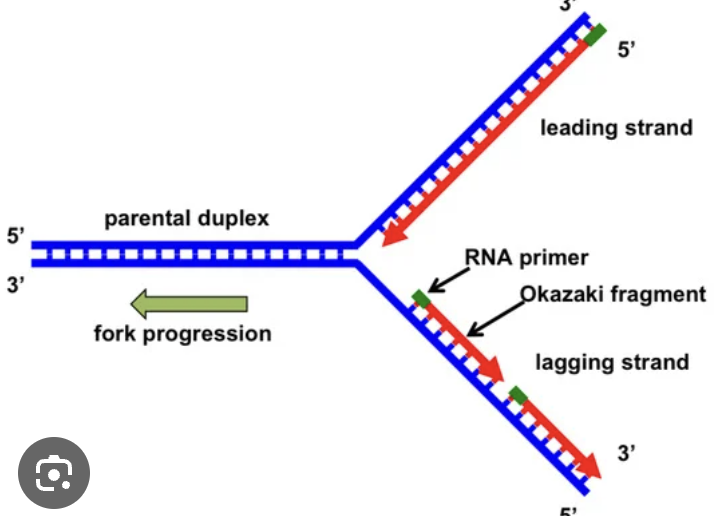
leading strand
5’ to 3’; synthesis occurs continuously
lagging strand
3’ to 5’; synthesis is discontinuous
Okazaki fragments
the result of synthesis occurring discontinuously on the lagging strand
DNA “nicks”
gaps of missing phosphodiester bonds resulting from the removal of RNA primers on fragmented lagging strands
DNA super-coiling
double helix twisted in space about its own axis; caused by the replication fork
proof-reading
action taken place in 3’ to 5’ direction by DNA Pol 1 and DNA Pol III
replisome
the complex of replication proteins that form at the replication fork; moves as a unit along the DNA
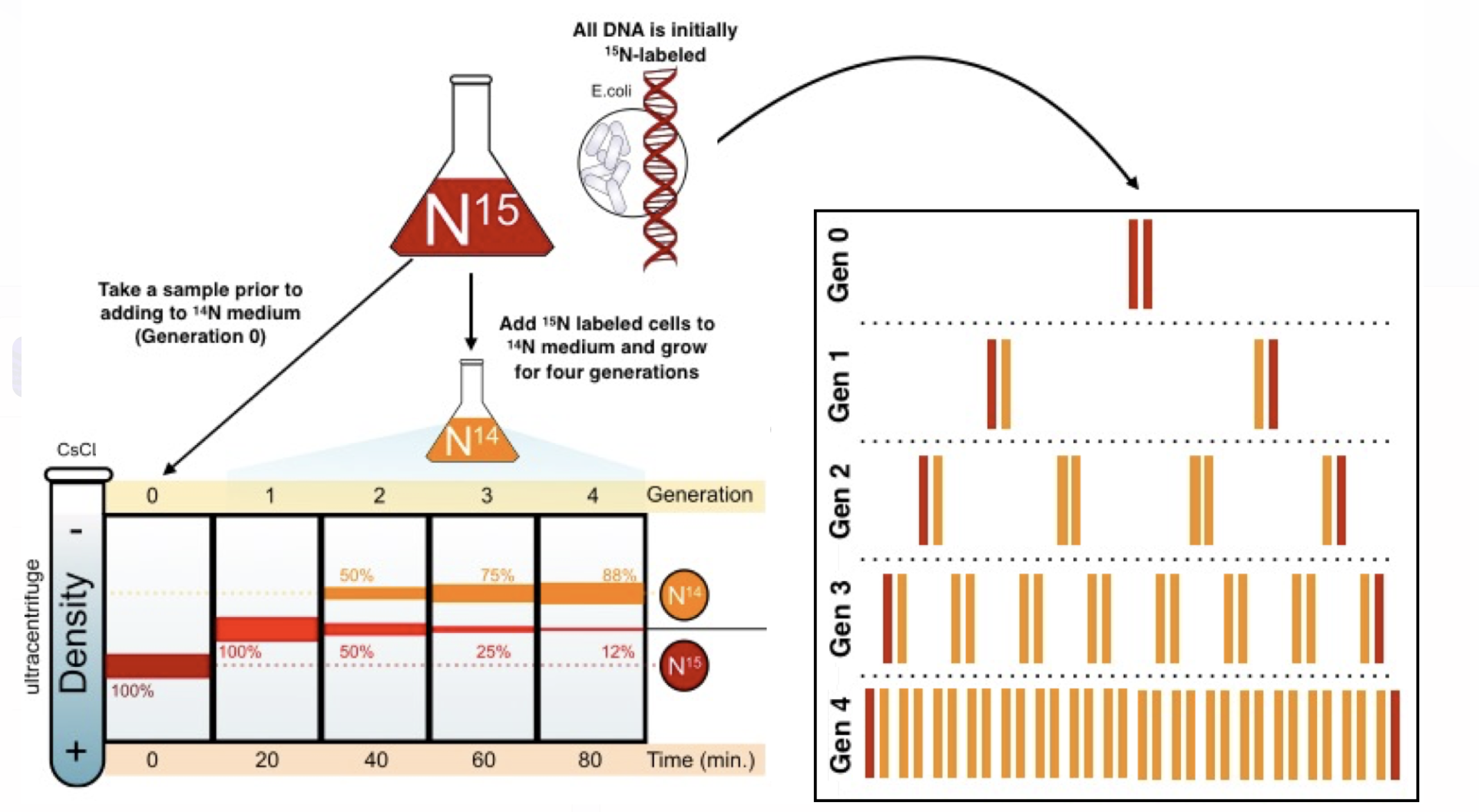
Meselson and Stahl’s experiment and conclusions
confirmed that DNA replicated through semi-conservative model; used heavier 15N isotope of E. coli DNA as well as a natural light 14N DNA media to measure how much of the 15N was in the sample after each replication
Kornberg’s experiment and conclusions
determined that the components necessary for DNA replication were the template, primer, DNA nucleotides (dNTPs) and DNA polymerase I
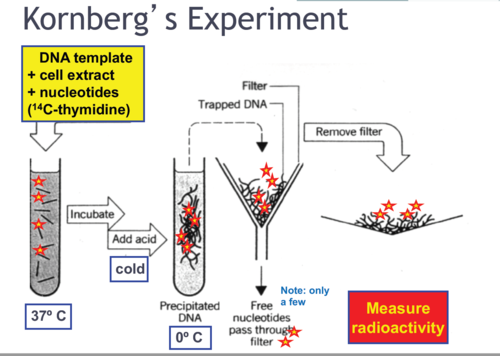
What components are needed for DNA replication in vitro?
template
primer
DNA nucleotides (dNTPs)
DNA polymerase + Mg2+ to increase efficiency of DNA pol
DNA replication in living cells
be familiar with replication fork and DNA synthesis on both strands
replication bubble
locally denatured segment of DNA
coding strand
sequence directly matches produced RNA
mRNAs
messenger RNAs that code for proteins; aka coding RNAs
tRNAs
transfer RNA; serves as a link between mRNA and growing chain of amino acids that make up a protein
rRNAs
ribosomal RNA; non-coding; acts as a catalyst to carry out protein synthesis
non-coding RNA
includes rRNA and tRNA as well as RNAs used for splicing / processing
RNA polymerase
capable of initiating new RNA chains (no primer needed)
RNA nucleotides
uracil, guanine, cytosine, adenine
uracil
RNA contains uracil in place of thymine; can form two bonds with A and G
GU “wobble” base-pairing
occurs due to 2 bonds with G instead of 3 (like in a typical GC pairing)
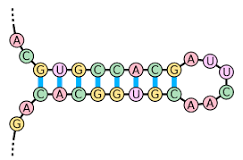
stem-loop RNA / hairpin
structure in RNA that forms when complementary sequences on the same strand pair up
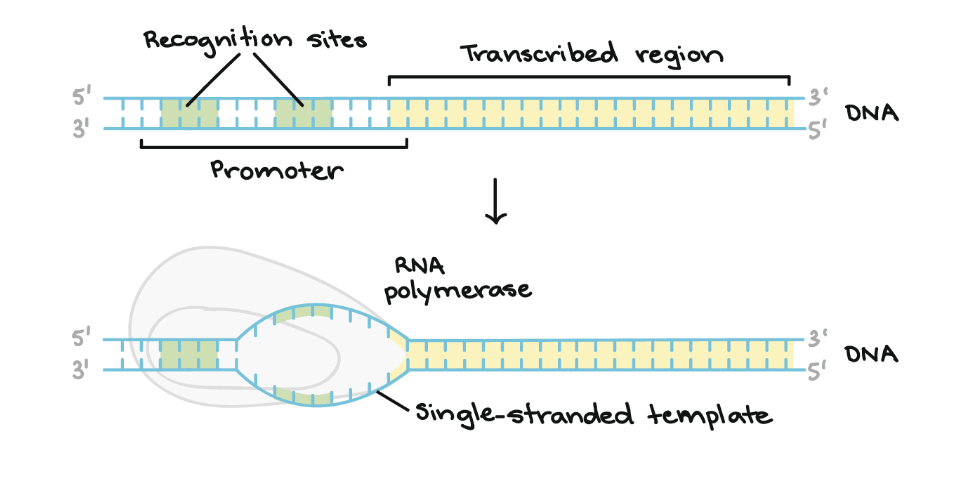
initiation of transcription in prokaryotes
formation of closed promoter complex
conversion of closed to open promoter complex
polymerizing the first 10 nucleotides while polymerase remains at promoter
promoter clearance (transcript becomes long enough to form a stable hybrid with template strand
elongation
after the first nucleotide, there is processive addition at growing 3’ end of RNA
termination in prokaryotes
two methods: Rho-independent, Rho-dependent
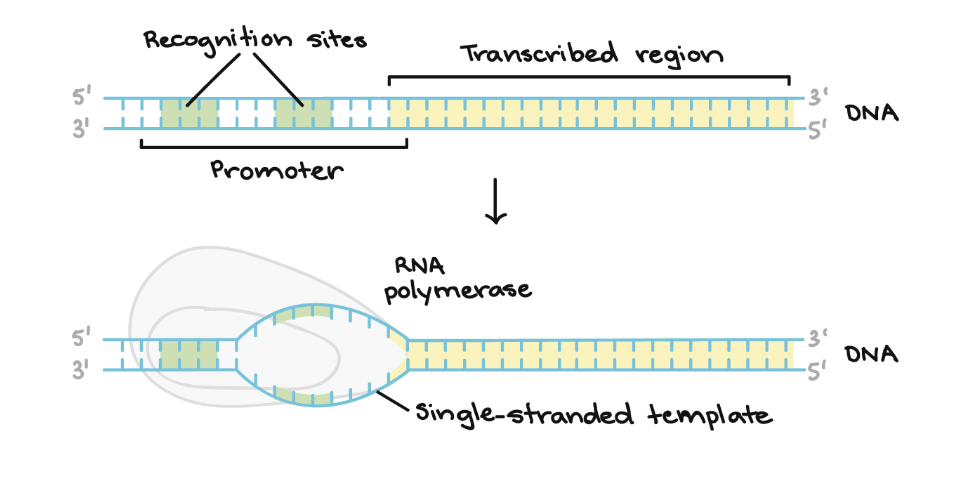
promoter region
the DNA sequence located upstream of a gene (-35 box); typically TA rich (weaker); helps RNA pol transcribe a gene
terminator
a section of nucleic acid that signals the end of the transcription process
upstream
numbers decrease (-35,-10 boxes)
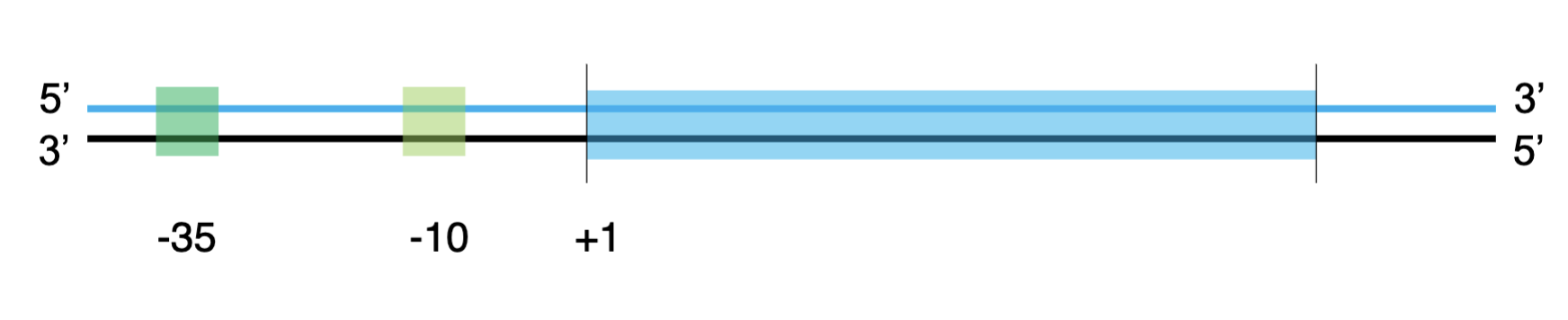
downstream
numbers increase
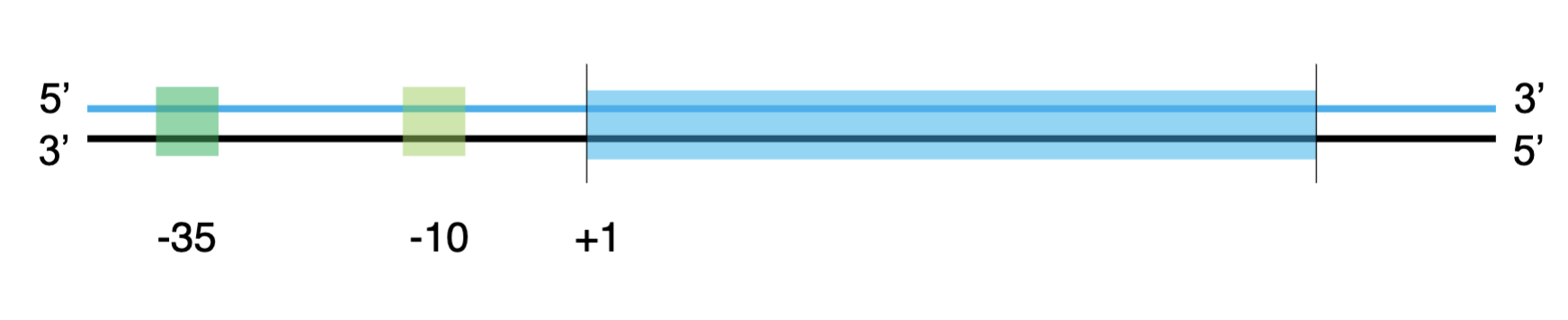
the +1 position
where the 1st nucleotide is transcribed
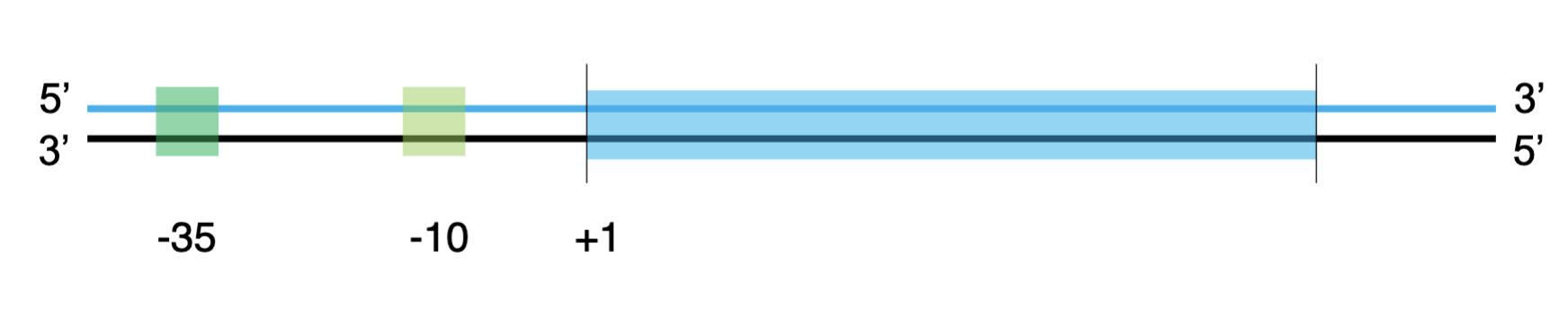
sigma factor
a protein; the sign post for RNA pol to initiate transcription
-10 box
part of the promoter located at base pair -10; where RNA pol begins to unwind DNA
-35 box
part of the promoter located at base pair -35; collaborates with Sigma factor to recruit RNA pol holoenzyme
open promoter
a DNA-RNA polymerase complex that’s partially unwound in preparation for transcription
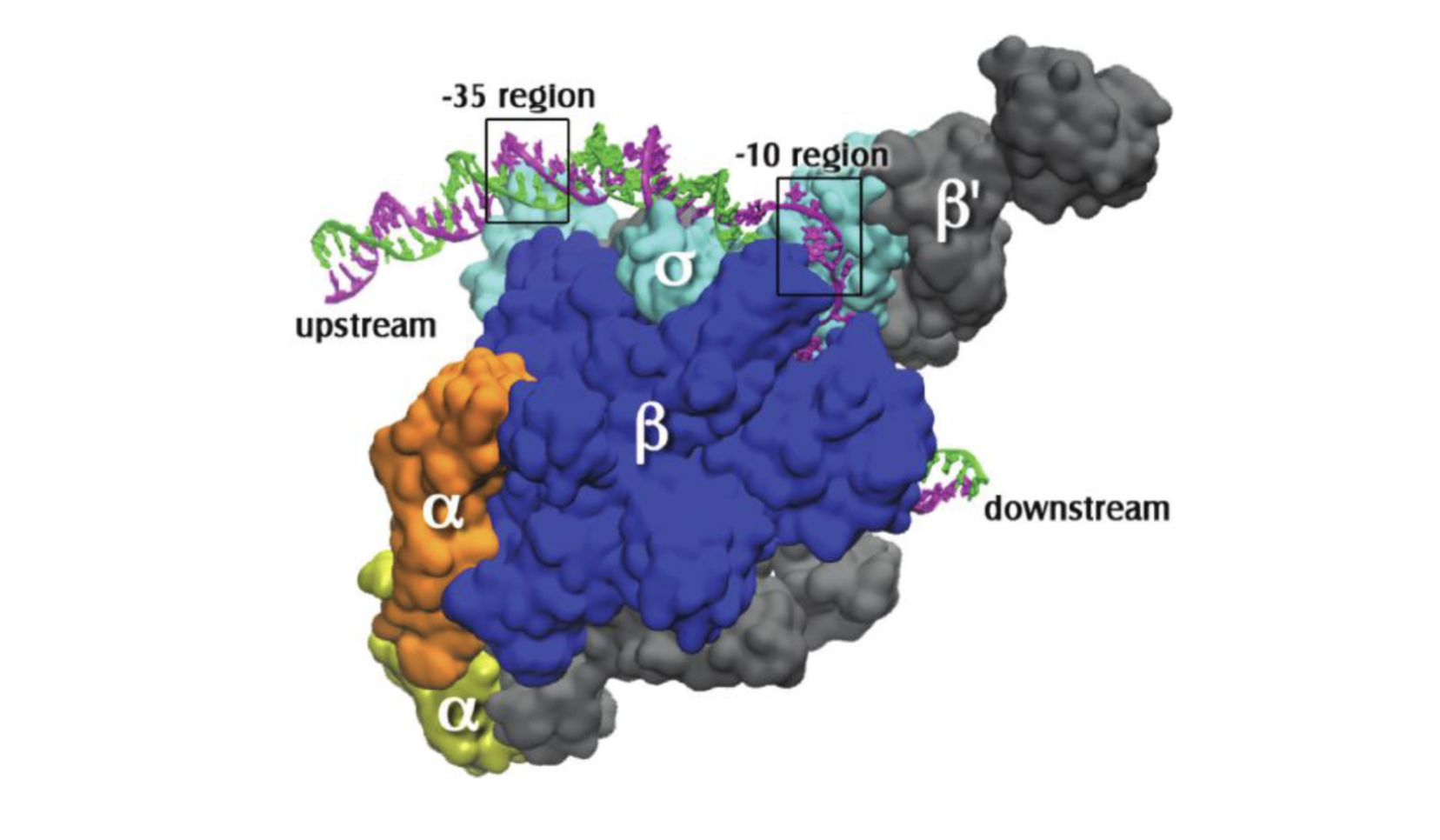
closed promoter
when RNA pol has bound itself to DNA that is still double-stranded
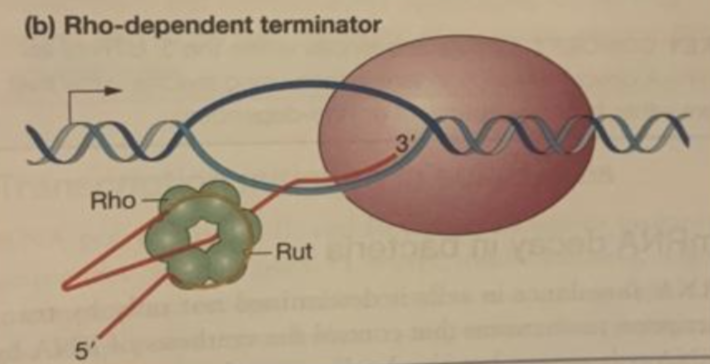
rho-dependent
when a rho protein binds to RNA, RNA pol dissociates from DNA and transcription ends
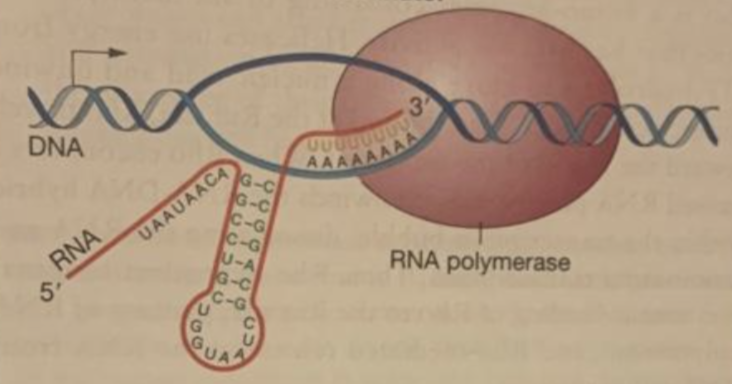
rho-independent
when a hairpin forms, RNA pol dissociates from DNA and transcription ends
base-pairing in RNA-RNA duplexes
NOT predictable