BIOL220 Exam 2
1/149
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
150 Terms
Summarize information flow using the central dogma and note any essential revisions and exceptions
DNA gets transcribed into mRNA, and mRNA gets transcribed into the protein
DNA can also get transcribed into ncRNA, which are exceptions to central dogma due to the fact it is RNA tha does not code for protein
What are some examples of ncRNA?
tRNA
rRNA
miRNA
How can RNA go back to DNA?
if it is a virus or we use reverse transcriptase
Wha else does RNA have beyond being a transient communicator?
It also has its own function based on structure
What is Dr. Zwemer’s definition of a gene?
A segment of DNA (includes promoter and proximal regulatory regions) that gets transcribed and has a function
Do we have a gene for each protein? How do we have multiple types of proteins?
We don’t have one gene for each protein but because of alternative splicing in eukaryotes we can generate multiple types of proteins from an individual gene
essential vs. non-essential amino acids?
essential amino acids we can build ourselves
non-essential amino acids we have to consume
what is ribonucleo protein complex for ribosome?
it is the combination of RNA and proteins (protein came from RNA)
what is rRNA?
rRNA is the central component of ribosomes
it is a ncRNA
it confers catalytic activity of ribosomes
What is special about proteins that are part of the ribosome?
these proteins are created using mRNA because they are a result of translation which is guided by mRNA
what is a linear sequence of DNA? what is linear sequence of amino acids
DNA: nucleic acid; polymer nucleotides connected by phosphodiester bonds
amino acids: polypeptide; peptide bonds link amino acids
for a given gene, which strand of DNA will be transcribed?
only the template strand will be transcribed; it is what RNA polymerase reads and produces reverse complementary of
What is the template strand of DNA?
3’ to 5’, so RNA sequence will begin 5’ to 3’
coding vs template strand of DNA vs strand of RNA?
coding gives same exact info as RNA bcs it is reverse complement to template strands (like RNA)
DNA has thymine and RNA has uracil
how does translation start in eukaryotes? prokaryotes?
eukaryotes: 5’ end of template strand enters the ribosome and ribosome scan for the first incidence of start codon (AUG)
prokaryotes: have a specific place to bind in ribosome called shine del garno sequence
How does ribosome set the frame?
there are no codons until the ribosome finds the first AUG relative to the 5’ end
how do you know if it is a prokaryotic or eukaryotic gene?
prokaryotic gene has -35 and -10 element
where does the promoter sequence lie relative to transcription start site?
it lies upstream, the 5’ direction relative to coding strand, also relative to what is transcribed
different genes have different promoter sequences but what do they share? (prokaryotes)
consensus sequence motifs are identical (which is the -10 and -35 elements in prokaryotes)
what happens if you change the nucleotide associated with elements?
the strength holoenzyme is able to bind to sigma factor is changed; the initiation efficiency of transcription is changed
what is a consensus sequence? do eukaryotes have consensus sequences?
consensus sequence are sequences that are conserved (in case of prokaryotes, -10 and -35 elements are consensus sequences bcs they are in all prokaryotic promoters)
eukaryotes have consensus sequences but it does not have perfect conservation; certain sequence motifs that will be there over and over again in the core promoter but they will not be a perfect conservation
what is one type of consensus sequence in eukaryotes? is it always in the same area? how does that affect expression?
core promoter elements are a type of consensus sequence and they are conserved
some core promoter elements are shared, but they are not always in the same area, this leads to a difference in expression because the then the polymerase will bind to different areas of the DNA sequence and transcribe different areas
cis-elements vs trans-factors?
cis: regions of DNA where trans-acting factors bind in a sequence specific manner
DNA sequences within or near to gene, which are recognized and bound by factors involved in regulating gene transcription
it cannot be moved
trans: originally encoded by gene so proteins and RNA
can be moved
what does translation in the cell?
ribosomes
what does transcription factors do? what is we mutate cis-element, how will that affect transcription factors?
it migrates back to the nucleus and binds to region of cis element (DNA) that is has special affinity to and influences the speed/strength of transcription
transcription factors are proteins so they are trans acting factors
mutation of cis element will alter how the transcription factor will bind to the DNA and potentially alter the regulation of transcription
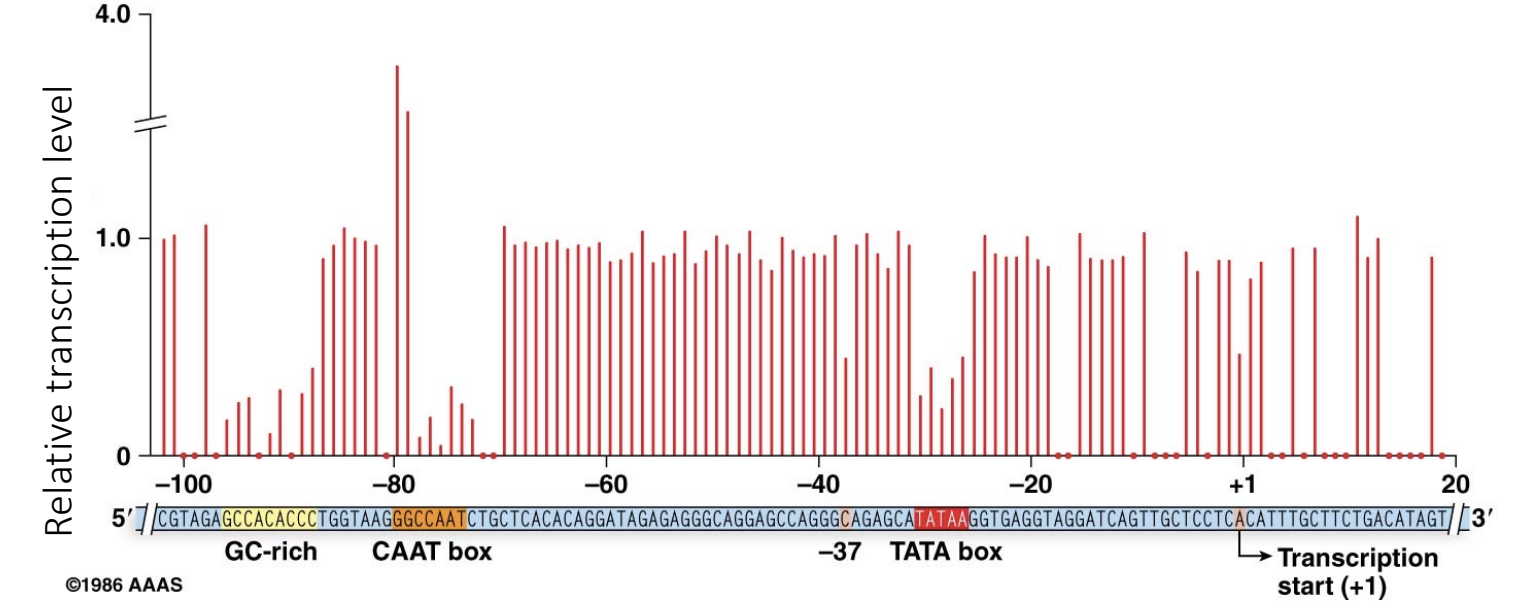
Explain this graph? Why is it important? What does it show?
scientists mutated various sites within the DNA sequence upstream of and surround promoter sequence of a gene, these sites are cis elements
impact of mutation based on how well transcription factor binds to them after mutation of cis element
each individual car is its own individual experiments and it tell you how transcription changes if a mutation was introduced at any one of the nucleotides
if RTL = 1 then a mutation at that nucleotide has no impact
if RTL < 1 then mutation had a big negative impact on amount of transcription produced/RNA produced
if RTL > 1 then mutation increases amount of transcription
Conclusion: some sequences in gene’s DNA can be mutated without affecting transcription efficacy (transcription level). For some sequences, however, a mutation would disrupt transcription (possibly affect overall organismal fitness). The sequences that cannot be mutated are usually evolutionary conserved and we call these conserved sequences “consensus sequences”
transcription can end up changing cellular function which can end up changing the phenotype of the organism
consensus sequences do not have a lot of variation (can include promoters, and specific transcription factor binding sites)
If promoter serves as cis acting element, in prokaryotes and eukaryotes, which factor(s) act(s) in trans
transcription factors in both prokaryotes and eukaryotes (more in eukaryotes)
sigma factor
any protein that is binding at cis element in a sequence specific manner
a way of modulating transcription are transcription factors
what trans acting factors have an effect on transcription?
transcription initiation
sigma factor (prokaryotes)
special transcription factors confirm tissue specificity (eukaryotes)
what is associated with every molecule?
energy
what are grooves in DNA? major groove vs minor groover?
the grooves of DNA are what proteins interact with
major grooves: indicates span that is the largest and offers more chemical information than minor groove
minor groove: indicates span that is the smallest
when looking at DNA form side, we can engage with more carbon and nitrogen atoms if coming along side the major groove
both are measured when looking at DNA sidon; unequal in width due to angle of bonds of nitrogenous bases attached to deoxyribose sugar
nitrogenous bases specific hydrogen donor/acceptor feature is info available to protein that is transversing major groove or minor groove
why is major groove better for protein binding than minor groove?
interaction based in H donor/acceptor provides more information if approaching from the major groove
exposed to more rich chemical energy landscape as a transacting factor at the major groove
allows for better specificity to ensure binding to unique sequence
what affect does adding a methyl group have to chemical landscame proteins encounter?
it affects the interaction between DNA binding proteins (transcriptional machinery & sequence)
how do we regulate transcriptional levels (prokaryotes)?
transcription initiation
how do we increase transcription level in prokaryotes?
faster and more tightly we bind RNA polymerase holoenzyme to promoter the more transcript will be produced
strength depends on match between the sigma factor and the cis element (promoter)
wha makes up the RNA polymerase of prokaryotes?
alpha, alpha, beta, beta prime, omega, and sigma factor
this is called the holoenzyme
how many RNA polymerase in prokaryote? what effect do sigma factors have on prokaryotes response to different environments?
just one RNA polymerase, but multiple different sigma factors which allows them to respond differently to environments and stressors
what sigma factor is majority of genes in bacteria transcribe to?
sigma 70
there are four other that regulate it
what are the three RNA polymerases of eukaryotes? what are their functions?
RNA polymerase I transcribes rRNA (in nucleus)
RNA polymerase II transcribes mRNA and snRNA (majority of transcription done for mRNA is done by RNA pol II)
RNA polymerase III transcribes tRNA and 5s rRNA
how do eukaryotes control how genes are expressed?
through controlling the availability and abundance of RNA polymerase II molecules; without RNA pol. II would mean it would be less likely for expression of genes it transcribes
what else does eukaryotic RNA polymerase need to transcribe?
basal transcription factors
TAF (TBP associated factors)
TBP (TATA binding protein)
TBP + TAF = TF2D (crucial transcription initiation factor)
RNA polymerase cannot associate with the promoter without basal transcription factors
what are the three stages of transcription?
initiation
elongation
termination
how do cells regulate gene expression?
regulation of transcription
more transcription means more proteins
less transcription means less proteins
what is the major way to regulate transcription? (prokaryotes)
initiation
depends on how well polymerase binds to gene promoter and allow it to engage in initiation process
what is the more abundant type of RNA in the cell by the # of different genes? # of molecules?
number of diff gene: RNA pol II
number of molecules: RNA pol I
what transcription factor do prokaryotes need for RNA polymerase to start going?
sigma factor
it binds to promoter in order for polymerase to bind to the promoter as well
how many consensus sequences do e. coli promoters have? what are they?
e. coli have two: positioned at -10 and -35 with respect to transcription start site
what is associated with transcription start site?
+1
what gives RNA polymerase holoenzyme its intrinsic helicase activity? why does it need to act as a helicase?
the subunits are what gives it i’s intrinsic helicase activity; needed because RNA polymerase has to get single strand of DNA to polymerize , add together RNA nucleotides that are reverse complementary to single strand of DNA; need to separare them because have a domain the holds strand not being used (non-template strand)
Describe what happens in initiation?
sigma factor binds to core promoters
RNA polymerase holoenzyme recognizes promoter at -35 region and binds to full promoter
RNA polymerase binds more tightly to the promoter at the -10 region, accompanied with local untwisting of DNA in the region
RNA polymerase correctly oriented to begin transcription at +1
what is another name for -10 element?
pribnow box
which is essential for transcription initation? what does the other element do?
-10 (pribnow box) is essential for transcription initiation
if deleted, there would be no transcription
-35 element allows for high level of transcription; it allows for better binding of polymerase enzyme to promoter
can be deleted the transcription just won’t be as good
how do we get better binding of sigma factor and polymerase?
the closer the promoter sequence matches the consensus sequence the better the binding
tight binding due to good chemical ineteraction between DNA and protein (specifically sigma factor)
if DNA binding pocket matches topological shape of DNA at cis-element where it is binding, the proteins is able to bind more tightly to the DNA
how do prokaryotes control expression?
through the use of different sigma factors and the abundance of them in the cell control the expression of genes needed under unusual circumstances
the -35 and -10 sequences are different for different sigma factors; changes which genes are expressed
how do we refer polymerase + sigma factor?
holoenzyme
what does transcription initiation depend on?
the sigma factor binding to the promoter; if it doesn’t bind the RNA polymerase cannot find the promoter.
what happens after ~10 nucleotides have been transcribed?
the polymerase continues to transcribe but the sigma factor falls off
after sigma factor fall off it is regulated to be removed in targeted way or inactivated in targeted way or else it can continue to bumble around and bind RNA polymerase again and restart transcription initiation
what marks the transition to the elongation phase?
the promoter clearance, the sigma factor falling off
Describe the formation of the pre-initiation complex of eukaryotes
TF2D (TBP and 13 other factors (TAF)) if helped by TF2A to bind a TATA box and other proximal core promoter sequences
TF2B joins and binds to additional sequence elements (BRE’s)
remaining basal transcription factors (E, F, H and mediator) bind as well and help recruit the RNA polymerase II
the complete PIC can now begin unwinding (done by TIF2H)
do eukaryotic RNA polymerase have intrinsic helicase activity?
no, we need basal transcription factors to provide it
what does the mediator transcription factor do?
mediates interaction of long range elements with promoter
which strand of DNA is bound by transcription factors?
both strands because most DNA binding proteins bind to the major groove, but the groove only exists when there is 2 strands of DNA
what acts as helicase in eukaryotic transcription initiation?
TF2H acts as helicase and unwinds dsDNA using energy supplied by ATP
What happens after PIC is in place?
TF2H acts as helicase and unwinds dsDNA
transcription bubble is formed and each strand of DNA must be stabilized within the polymerase but only the template strand is used to produced RNA
first 10 ribonucleotides synthesized and leave via the exit channel
promoter escape and we enter elongation phase
can different strands of the double helix DNA be used as template for different genes?
yes, it depends on how the DNA is being read; polymerase always read 3’ to 5’ and build 5’ to 3’
what happens once polymerase moves past the template and non-template strand?
they will spontaneously reanneal
prokaryotes vs eukaryotes in transcription, translation mechanisms?
prokaryotes: co transcription translation because there is no nucleus, and transcription happens in cytosol which is where translation also occurs
eukaryotes: no co transcription translation because RNA is in the nucleus and has to be transported out of the nucleus to be translated; won’t be exported out until it is processed
processing both co transcriptional (5’ capping and intron splicing) and after cleavage of the transcript (addition of poly A tail)
what is termination?
it is the dissociation of RNA polymerase, RNA and DNA from one another
describe intrinsic termination
rho independent
one of major focuses is formation of secondary structure of RNA due to self complementary
creates a hairpin, it is reverse complementary to itself and hybridizes (top does no hybridize)
relies on transcription termination sequence, which is a part of the gene and also gets transcribed
while RNA polymerase is transcribing it gets to a point where the RNA it creates makes a stem loop or hairpin, where it hybridizes to itself, in which a NusA protein binds to it and causes the polymerase to temporarily stall. so now the only thing keeping DNA and RNA intact is the poly U tract which has really weak bonds due to only having two hydrogen bonds. bcs of this weakness, it will just dissociate and the polymerase will fall off and transcription ends
describe rho dependent termination (extrinsic)
depends on rho protein
relies on presence of rho binding site sequence getting transcribed into the RNA as well as the presence of a transcription stop point within DNA (not transcribed) to stall RNA polymerase
rho binding site sequence has to be transcribed
rho protein binds to RNA at reverse complement of that sequence
transcription stop point is located at certain distance after rho binding site
As RNA polymerase transcribes the DNA, it comes across a Rho binding site sequence, which it transcribes into RNA. The Rho protein binds to this site on the RNA and climbs along the RNA, using ATP to move toward the RNA polymerase while spinning through the RNA strand. When RNA polymerase reaches a termination site, it temporarily stalls, allowing Rho to catch up. Once Rho reaches the transcription complex, it uses its helicase activity to unwind the RNA–DNA hybrid, releasing the RNA transcript. The RNA polymerase then dissociates from the DNA, and transcription ends.
wha is the rho protein?
it is a helicase that climbs up RNA and unwinds the RNA DNA duplex to release the nascent transcript
it spins the RNA which decouples the RNA from DNA inside the transcription bubble
does polymerase continue polymerizing after gene is released?
yes
what must be completed in eukaryotes in order for RNA to be exported from the nucleus for translation in cytosol?
pre-RNA must be processed to become mRNA
it is both co transcriptional (5’ capping and intron splicing) and pos transcriptional (poly A tail)
why do we need 5’ cap and poly A tail?
it is needed to protect the RNA, without it the molecule would be digested by enzymes in the cytoplasm and little translation would occur
is splicing both co and post transcriptional?
yes because the cell is multi tasking
describe the 5’ cap, why is it important?
the 5’ cap is a nucleotide, it is a type of guanine
it prevents the degradation of mRNA
increases the efficiency of splicing of mRNA
necessary for exporting the mRNA out of the nucleus
allows the recognition of mRNAs by the protein synthesis machinery in the cytoplasm (enhances translation of mRNA
what is special about the 5’ cap?
it binds to the pre mRNA through a triphosphate bridge which connects 5’ to 5’
it is linked backwards
describe poly A tail addition, why is it important?
poly A polymerase adds As to 3’ end ~250 times
the longer the tail length the high the half life
prevents degradation of mRNA
associated with terminating transcription
necessary for transporting mRNAs out of the nucleus
optional assist in efficiency of binding to the ribosome for translation
why is there no real eukaryotic “termination sequence” in the case of RNA Pol II and DNA?
RNA polymerase transcribes right through the poly A tail sequence
describe the cleavage and polyadenylation executed by trans acting factors
it is executed in response to polyadenylation signal sequence transcribed into the pre mRNA
CPSF recognizes and binds cis-elements and catalyzes cleavage of RNA; binds to AAUAAA sequence
CstF lends additional specificity to binding of complex
polyadenylate polymerase builds poly A tail (~250 nucleotides long)
is eukaryotic mRNA cleaved and rleeased before RNA polymerase stops?
yes
can exons be shuffled?
no they cannot be shuffled, just included or excluded
what allows for hybridization? where would denatured gDNA and mRNA successfully hybridize? where would they no hybridize?
reverse complementary allows for hybridization
hybridize in UTRs
exonic sequences form gDNA will be reverse complementary to majority of mRNA
regions complementary to template strand of DNA
don’t hybridize in poly A tail and introns
regions where mRNA does not have complementary sequence
what does the r looping experiment show?
shows components of DNA are reflected in mRNA but some are not; introduced introns that use to be known as SPLIT genes (DNA split in segments included and excluded from mRNA)
what occurs in the lab relating to introns?
they are not being spliced and we have gDNA and mRNA to hybridize
what are snRNPs?
snRNA + proteins
what are spliceosomes? what is its function?
it is a complex snRNPs and other proteins that splices introns out of pre mRNA
in the nucleus, its function is to recognize introns and remove them and ligate exons together because we don’t want holes in the mRNA
what catalyzes splicing reaction?
protein component
what is he role of snRNA?
recognition of intron/exon boundary
why does the inclusion/exclusion of exons lead to different proteins?
each exon encodes for certain sequences of polypeptide and when the polypeptide is folded into protein structure, creates a protein domain with different structural features which allows for proteins to have different functions
how does spliceosome recognize crucial sites?
it recognizes through conserved sequences and sequence milieu
recognition depends not only on specific nucleotides (splice site donor/splice site acceptor) but also on which nucleotides are nearby
where is the splice donor site? splice acceptor site?
donor is at the 5’ beginning of an intron
acceptor is at 3’ end of intron
explain the splicing mechanism
a greedy adenosine with a 2’ OH executes an attack at the intron/exon boundary
bond is formed between 2’ hydroxyl and 5’ guanine (attack splice site donor)
forms a lariat
3’ end exon I has free hydroxyl group so spliceosome assists 3’ hydroxyl to attack at other introns/exons boundary
3’ of exon I forms a phosphodiester bond with 5’ carbon of beginning of exon 2
product is spliced exonic sequence and excised lariat
what would happen if there is a splicing mutation?
there would be a difference in the exons that are ligated together, which would cause a difference in what protein is produced.
what gene does hemophilia occur in?
F8 or F9
what does it mean that alexai was hemizygous when diagnosed with hemophilia?
he only had one allele present due to hemophilia being a X-linked gene
what does recessive mean?
absence of dominate allele with at least one copy of recessive allele
explain the cryptic splice acceptor site
here was a mutation that caused the splice site acceptor to be read earlier in the sequence causing a frame shift
the spliceosome gets confused on which one is the splice site donor
describe the initiation of translation in eukaryotes
translation starts with assembling of small subunit of the ribosome, the initiator tRNA and the initiation factors at the 5’ cap.
pre initiation complex then slides downstream until it reaches the codon AUG, setting the frame
initiation complex is finalized with addition of large subunit of ribosome
the 5’ end enters the ribosome and ribsome reads 5’ to 3’
describe translation of mRNA in eukaryotes
mRNA enters into small subunit and large subunit attaches after it
initiator tRNA positioned at P site and tRNA at A sie
whichever tRNA is in the P site is called the peptidyl tRNA because it is the one that holds the growing chains of polypeptides
each amino acid has an ester bond that connects it to the tRNA that is cleaved so the peptide bond can be formed between the amino acid and the next amino acid
ribosome stops holding the tRNA that no longer has amino acid
release factor enters A site and causes polypeptide chain to be cleaved from tRNA in P site to be released
what are the three steps of translation?
initiation is the assembly of ribosome around mRNA and first charged tRNA
elongation is peptide bonds forming as charged tRNAs bring appropriate amino acids to ribosome
termination is stop codons releasing factors and complex dissociates