Introduction, Regulatory Proteins, & Protein Binding Properties
1/81
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
82 Terms
objectives and themes of this course
A. Learning the basic physiological principles that direct the activity of the human body.
B. Determining what is “normal” for the body – what homeostasis is and what homeostasis is not.
C. Understanding the connectedness between physiological system and applying the underlying influences of chemistry, physics, and (relatively simple) mathematics.
D. Understanding how pathology occurs, and why diseases create the symptoms we observe.
why are humans special from machines even though there are some that can perform multiple functions?
our bodies are able to perform many diverse functions, but any one of the machines we make is typically able to perform only one or a few of these same functions
what allows our body to perform various functions?
electrochemical interactions
electrochemical interactions
interactions between molecules that occur because of the specific electrical or chemical properties of the molecules involved, and due to the electrical and chemical properties in the surrounding environment (cell cytoplasm, extracellular fluids, etc.)
what are electrochemical interactions controlled by?
many regulatory molecules
regulatory molecules
these are molecules that possess binding sites that temporarily and weakly bind specific molecules (ligands), forcing the molecules to change or interact with other molecules
most regulatory molecules are ____
proteins
what do proteins control?
a vast variety of chemical reactions in the body
why are proteins able to control a vast variety of chemical reactions in the body?
our genome allows us to construct a great diversity of proteins, each molecularly and physically distinctive
what are some examples of regulatory proteins?
enzymes, transcription factors, structural molecules, binding proteins, hormones & other chemical messengers, receptors, membrane pumps and channels
a protein’s properties are determined by what?
by its shape
primary (1°) structure
the amino acid sequence of the protein, formed by covalent peptide bonds
secondary (2°) structure
specific α-helix or β-pleated sheet folds of the protein, formed by H-bonds
tertiary (3°) structure
all other folds or bends in the protein, formed by various types of bonds, mostly weak non-covalent (hydrogen- and ionic-) bonds, but sometimes by covalent disulfide bonds
quaternary (4°) structure
non-covalent association of two or more polypeptide subunits to form one functional protein
what determines the properties of a protein’s binding site?
the final shape of a protein
binding site specificity
a measure of how many different ligands are able to fit into one binding site
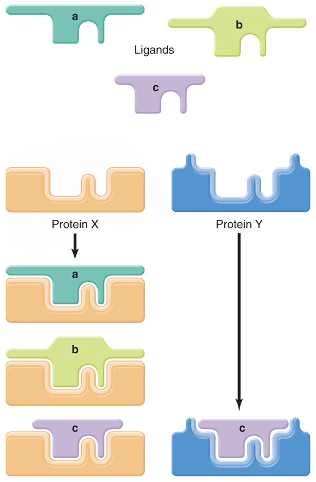
low specificity
many ligands can bind
e.g., hydroxysteroid dehydrogenase enzymes-binds many steroids
high specificity
only binds a few molecules
e.g., hemoglobin-binds O2 and CO (not CO2) well at the O2 site
binding site affinity
a measure of how strongly the binding site binds a ligand
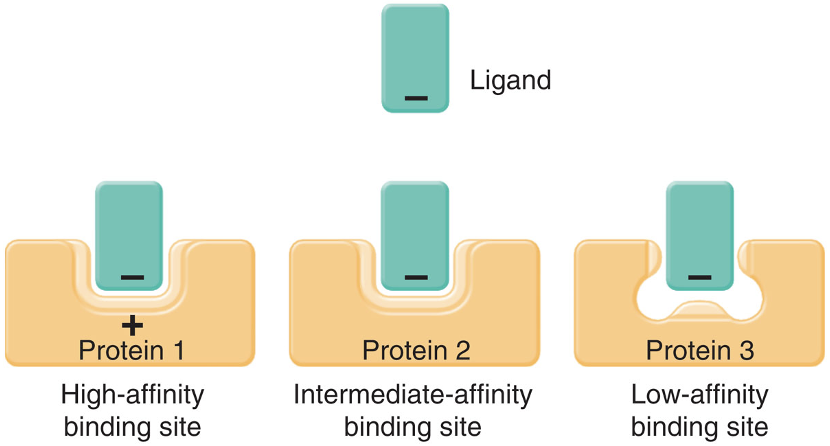
low affinity
there is weak binding between a molecule (ligand) and its target (receptor or protein), requiring a higher concentration of the ligand to achieve effective binding
high affinity
there are strong attractive forces between a ligand and its target, resulting in a stable complex and a sustained response
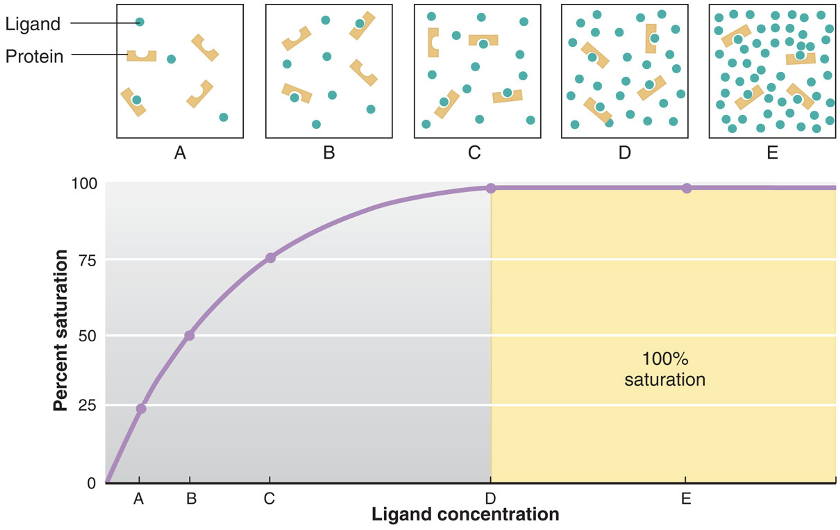
binding site capacity
a measure of how many binding sites present in a cell or tissue sample
binding site saturation
a relative measure of how many binding sites are occupied at any moment (percentage value)
when all the binding sites are occupied, the population of binding sites is 100% saturated
when half the available sites are occupies, the system is 50% saturated
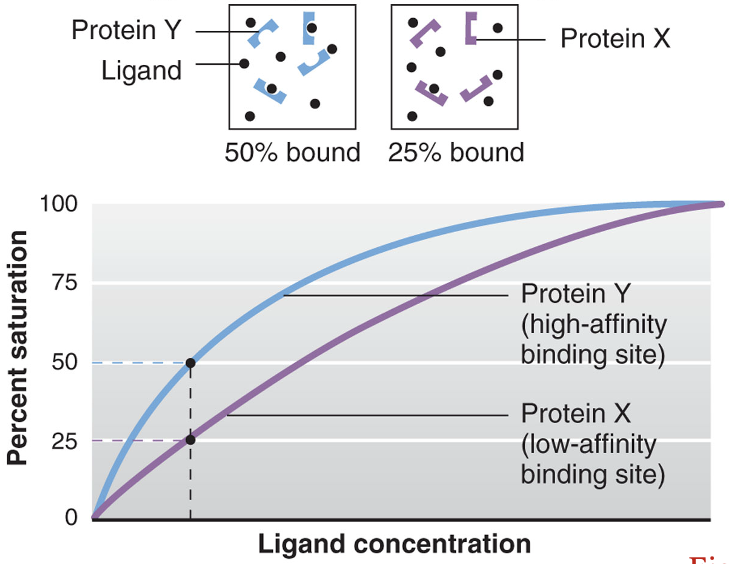
ligand competition
occurs when two different ligands compete for one binding site
the relative affinities and concentrations of the ligands determine which ligand occupies more binding sites
the affinity of Hb for CO is 250 times greater than the affinity of Hb for O2
if protein shape is change what can happen?
binding site affinity can be affected (changed)
what environmental factors can affect binding site affinity?
changing pH, temperature, osmolarity can alter the non-covalent bonds in protein
what molecular factors can affect binding site affinity?
allosteric interactions (non-covalent)
covalent interactions (covalent)
allosteric interactions
involve a modulator molecule binding to a regulatory site, which then alters the affinity of another binding site on the protein (the functional/catalytic site) for its ligands
covalent interactions
involve the covalent addition or removal of some chemical group (usually an inorganic phosphate group) to/from the regulatory protein, thus changing the affinity of the protein’s functional site/catalytic site for its ligand
denaturation
reversible or irreversible destruction of binding site activity by altering protein shape
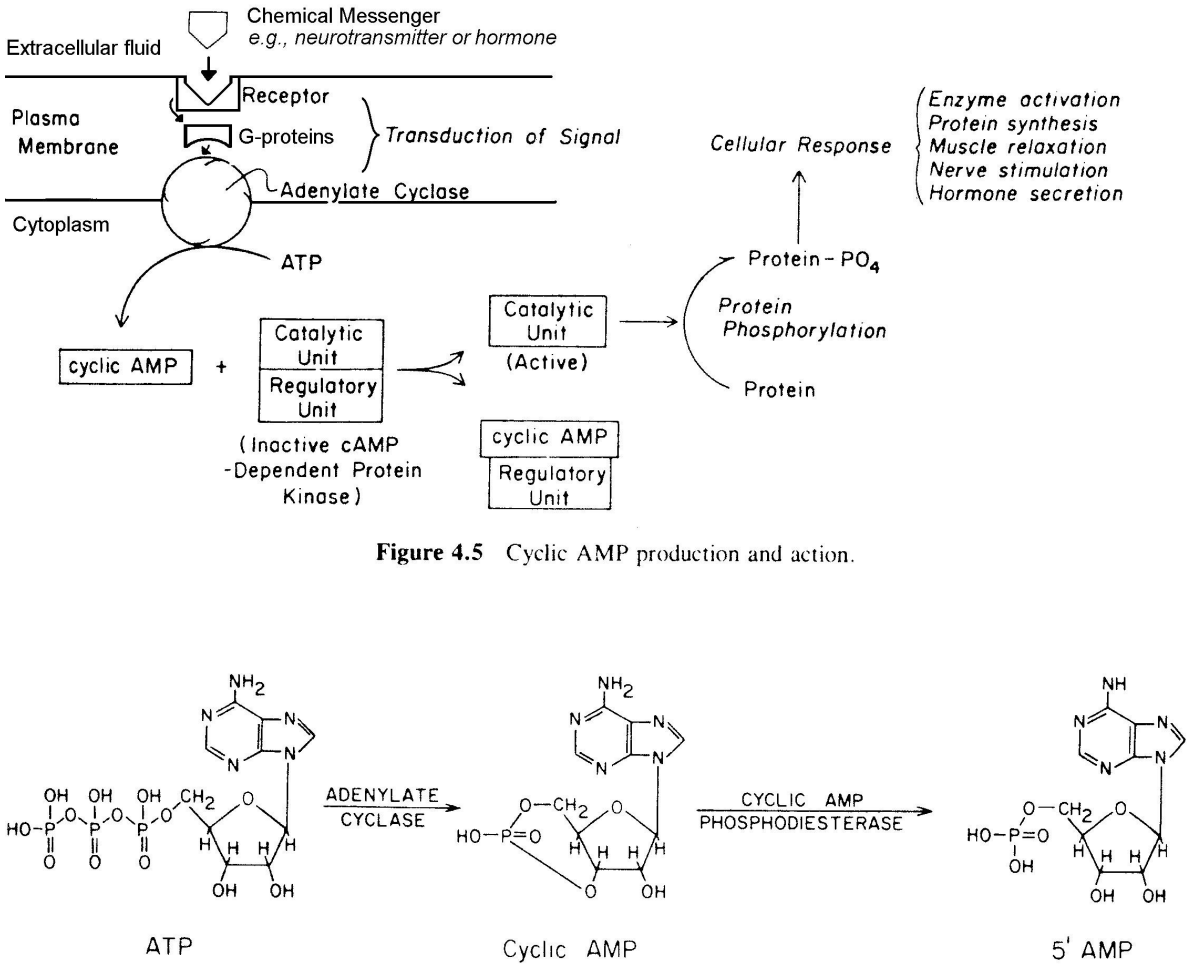
example of signal transduction form chemical messenger to cAMP to enzymes
this pathway is used by many cells and chemical messengers to alter cell physiology
it involves a combination of allosteric and covalent interactions:
allosteric changes are easily reversible
covalent changes are created when protein kinase enzymes add phosphates to proteins, and reversed when phosphoprotein phosphatase enzymes remove the same phosphate groups
what type of physiological activities does the body perform?
muscle contraction/movement
secretion
chemical reactions
electrochemical pumps
cell division
why do regulatory molecules have to be unique?
don’t want to turn on/turn off multiple processes
exception: when you want multiple processes to occur together
examples of regulatory proteins
molecular motors (e.g., actin/myosin)
enzymes
transcription factors
chemical signals (hormones, neurotransmitters, etc)
membrane channels
ion and solute pumps
what is the diversity of the reactions controlled dependent on?
diversity of regulatory proteins being available
protein diversity is the result of what?
variations in protein structure
what bond connects amino acids together?
peptide bond
N-terminus
amino group on the amino acid
C-terminus
carboxylic acid on the amino acid
which matters more: property or identity of the amino acid?
property
how are two amino acids connected?
dehydration synthesis
what variables determine the primary structure of a protein?
the number of amino acids in the chain
the specific type of amino acid at each position along the chain
what is a polypeptide in the primary structure analogous (comparable in certain respects) to?
a linear string of beads, each bead representing one amino acid
conformation
the final shape of a protein
based on interactions between side groups of each amino acid lead to bending, twisting, and folding of the chain into a more compact structure
native conformation
what is the shape of a protein where it belongs/the place it should be
compare the strength of hydrogen bonds and ionic bonds
they are about the same because ionic is weak in water and break apart once wet
strongest side chain interaction
covalent (disulfide) bond
alpha helix
because peptide bonds occur at regular intervals along a polypeptide chain, the hydrogen bonds between them tend to force the chain into this coiled conformation
beta pleated sheet
hydrogen bonds can also form between peptide bonds when extended regions of a polypeptide chain run approximately parallel to each other, forming a relatively straight, extended region
final three-dimensional conformation
once secondary structure has been formed, associations between additional amino acid side chains become possible
these interactions fold the polypeptide, making it a functional protein
random coil conformations
the sizes of the side chains and the presence of ionic bonds between side chains with opposite charges can interfere with the repetitive hydrogen bonding required to produce these alpha and beta shapes and result in irregular regions
when do final three-dimensional conformation and random coil conformations occur?
these occur in regions linking the more regular helical and beta pleated sheet patterns
what ability do beta pleated sheets and alpha helices impart upon a protein?
the ability to anchor itself into a lipid bilayer
domain
a part of a polypeptide chain
where is the amino end in terms of a membrane?
outside
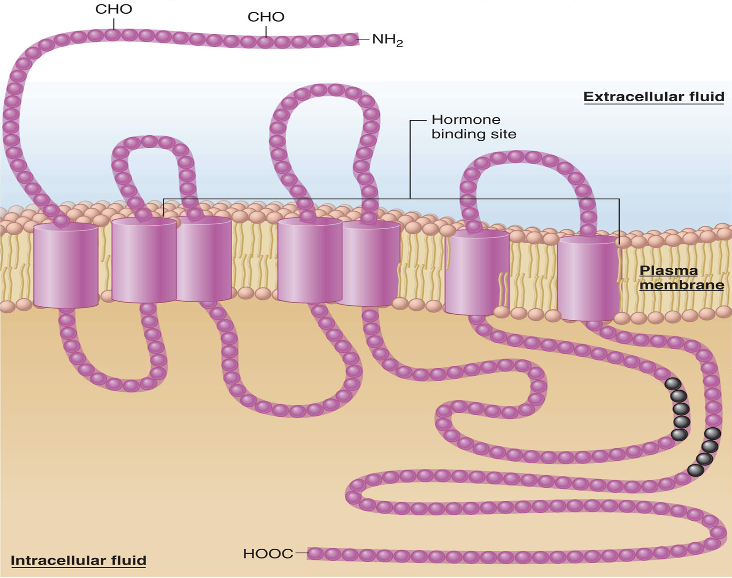
where is the carboxyl end in terms of a membrane?
in the cytoplasm
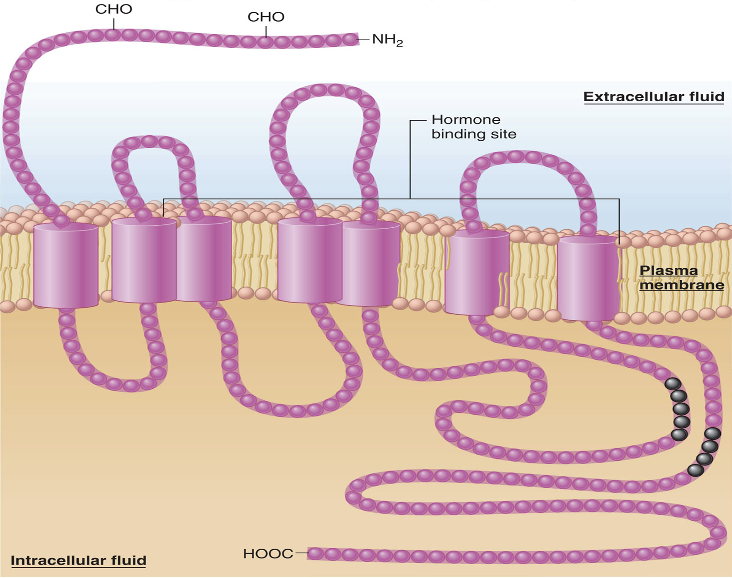
multimeric (“many parts”) proteins
name for proteins that are composed of more than one polypeptide chain and are said to have a quarternary structure
peptide
for a relatively short amino acid sequence (<40 aa)
polypeptide
a single chain that’s longer (>40 aa)
protein
could be used in place of peptide or polypeptide; some proteins have more than one polypeptide
what interactions form quarternary structures?
same as secondary structure (the chains can be held together by interactions between various ionized, polar, and nonpolar side chains, as well as by disulfide covalent bonds between the chains
the same factors that influence the conformation of a single polypeptide also determine the interactions between the polypeptides in a multimeric protein
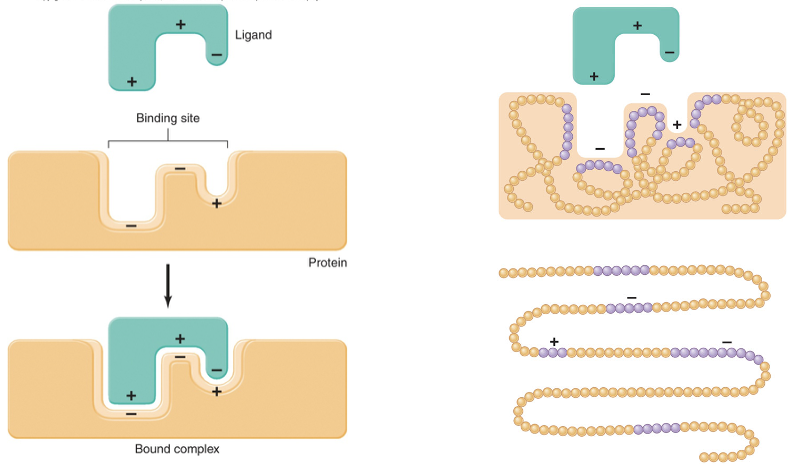
ligand
any molecule or ion that is bound to a protein by one of the following forces
electrical attractions between oppositely charged ionic or polarized groups on the ligand and the protein
weaker attractions due to hydrophobic forces between nonpolar regions on the two molecules
when a ligand bonds to a protein what type of bonds are not involved?
covalent
is the binding of a ligand reversible or irreversible?
generally reversible
binding site
the region of a protein to which a ligand binds
a protein may contain ____
several binding sites, each specific for a particular ligand, or it may have multiple binding sites for the same ligand
what is the usual result of the binding of a ligand to a protein?
the conformation of the protein changes
when this happens, the proteins’ specific function may either be activated or inhibited, depending on the ligand
lock and key binding
what does the percent saturation of a binding site depend on?
the concentration of unbound ligand in the solution
the affinity of the binding site for the ligand
at 0 K what happens?
every atom stops moving
brownian motion
the random movement of visible particles suspended in a fluid (liquid or gas), caused by the constant bombardment of the invisible, surrounding molecules of the fluid
any motion that occurs at > 0 K
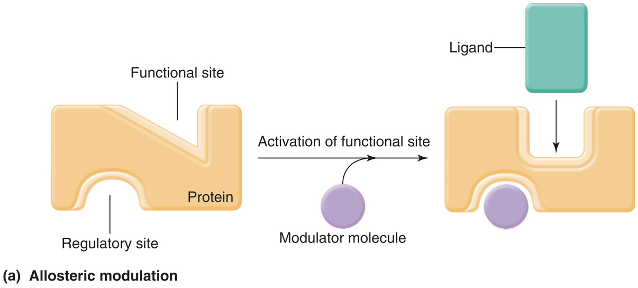
allosteric modulation
occurs when a protein has two binding sites and the binding of a protein to one of the sites alters the shape of the other
more quickly reversible
depend on the availability of the modulator
functional (or active) site
carries out the protein’s physiological function
regulatory site
the ligand that binds to the regulatory site is known as a modulator molecule, because its binding allosterically modulates the shape, and therefore the activity, of the functional site
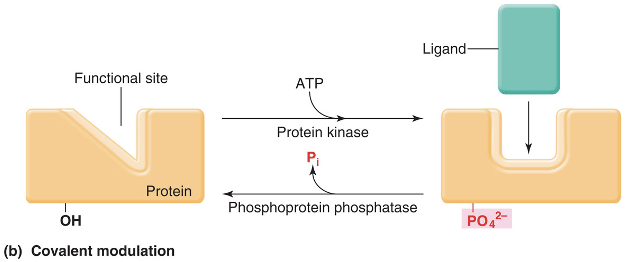
covalent modulation
is the covalent binding of charged chemical groups to some of the protein’s side chains
phosphorylation
the most common type of covalent modulation
addition of a phosphate group
require kinases
dephosphorylation
removal of a phosphate group
require phosphatases
kinase
an enzyme that adds the phosphate group to another protein
phosphatase
an enzyme that removes the phosphate group from a protein
how to kinases and phosphatases work?
they recognize specific sequences in the protein to find thier targets
second messenger system
relays extracellular signals, like hormones, to trigger intracellular responses
These systems involve small, intracellular molecules called second messengers (such as cAMP, calcium, IP3, and DAG) that amplify and propagate the signal, leading to changes in protein activity, gene expression, and other cellular functions.
The system amplifies the signal by generating many second messengers from a single first messenger, providing speed and flexibility for cellular responses