Genetics and biochemistry semester 2
1/109
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
110 Terms
what are the 2 types of mutations.
Somatic mutations (mutations of somatic tissues/body cells), and germ line mutations (mutations in gametes)
How can mutations be passed on to future generations.
Mutations can be passed to the daughter cells through mitosis. Germ-line mutations can be passed to future generations through fertilisation using mutated gametes. The offspring will carry that mutation along their somatic and germ line mutations.
Why do most somatic mutations have no obvious effects.
The function of the mutated cell is taken over by another cell in some cases.
The mutated cell dies and it is then replaced by a normal one.
How do cancers arise.
Mutations are found in cells that stimulate cell division. These cells have an advantage, the mutation spreads rapidly which causes the cancer to develop.
what are Proto oncogens
They are normal genes that play a crucial role in regulating cell growth, division, and differentiation. They are essential for normal development and maintenance of tissues in the body. They produce proteins that help cells grow and divide in a controlled manner.
What can a mutated proto oncogen cause. What is a term used to describe this mutated version.
Mutated proto-oncogenes can cause an increase in the production of a modified protein which stimulate cell division, differentiation and it can inhibit cell death. This can arise in cancer cells. Mutated versions are called oncogenes.
What are tumour repressor genes.
They are segments of DNA that code for negative regulator proteins which prevents unontrolled cell division. They can put a road block to cell cycle progression until certain events are completed.
What are the types of mutations caused by DNA polymerase errors.
Base substitutions, Insertion and deletoins of bases, expanding nuceotide repeats.
Explain base substitution mutations. What are transitions vs transversions in base substitution.
When the incorrect base is placed in the sequence. When the single codon is altered, it can change the type of protein it codes for. Transitions is when purines are replaced by purines (same same for pyrimidines) and transversions is when purine is replaced by pyrimidine vise versa.
What are the 3 types of mutations and what can be observed
Neutral mutation: alters the amino acid sequence but not the protein function.
Loss of function mutation: the protein is no longer functional
Gain of function mutation: The cell produces a new protein product which is often dominant.
What is a well known example for a disease which is caused by base substitution mutation. How does this occur
Sickle cell disease. A base substitution occurs on the gene encoding for Haemoglobin which changes glutamic acid into valine. This changes the shape of the haemoglobin, which impairs function.
Explain Inserion and deletions in terms of Mutation.
There are 2 ways it occurs. An extra nucleotide is added or is missing. It causes a shift in the reading frame which changes the type of protein it codes for. This is known as frameshift mutation. Another type is nonsense mutations. This happens when a base is taken out which creates a premature stop codon. This alters the protein shape. Cystic fibrosis is an illness caused by this type.
Explain expanding nucleotide repeats in mutations. Provide an example of an illness caused by this
It happens when the number of sets of repeated sequences increases within a gene. An illness caused by this is fragile x syndrome where there are repeats of the FMR-1 gene.
How does expanding nucleotide repeats cause Huntington disease.
There are repeats in CAG in the gene which produces a toxic protein that damages neurones.
What are some examples of diseases caused by expanding nucleotide repeats.
ALS, Huntingdons disease, Fragile X syndrome.
What are some examples of mutagens both physical and chemical.
Radiation: ionising radiation such as X-rays and gamma rays.
Radiation: non-ionising radiation such as UV light
Chemical agents: Base-modifying and intercalating chemical agents.
How does ionising radiation cause mutations.
X-rays penetrate tissues and knock electrons out of orbits and create ions. These ions can break covalent bonds in the sugar phosphate backbone, which results in single and double-stranded DNA breaks. This causes interchanges between chromosomes. Ionising radiation kills cells at higher doses and at lower doses it creaes these point mutations.
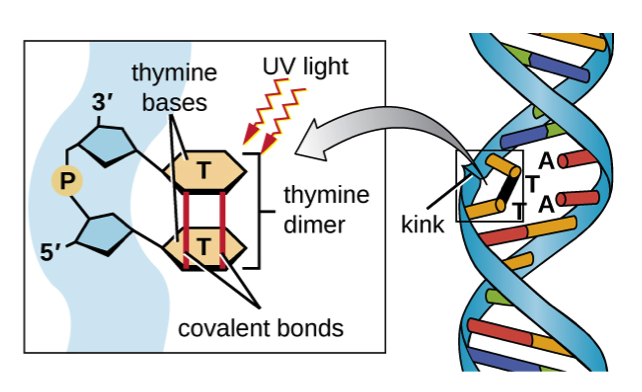
How does non-ionising radiation cause mutations.
UV is absorbed by purines and pyrimidines which creates abnormal chemical covalent bonds between bases. This is known as dimer. This can result in bulky lesions in the DNA that stall replication and transcription and introduces frameshifts or point mutations.

How do chemical agents cause mutations. Provide examples.
Chemicals called nucleoside analogues are structurally similar to regular nucleotide bases and can be incorporated into DNA during replication.
These base analogues induce mutations because they often have different base-pairing rules than the bases they replace. Introduce mutations during replication.
Examples include, 2-aminopurine nucleoside, acycloguanosine, and 5’-bromouracil.
What is EMS and why is it preferred over nitric acid to mutate organisms
EMS (Ethyl Methanesulfonate) is a chemical mutagen used to cause mutations in DNA.
EMS is not volatile (so it is less likely to be breathed in).
It is soluble in water (this solution then becomes very mutagenic).
It can be left overnight, and it becomes completely inactive; it can be disposed of safely.
What is crispr cas 9. What does it stand for?
It is a technology that allows for the editing of genomes and gene modification. It stands for clustered regularly interspaced short palindromic repeats.
What makes up the locus of CRISPR
Cas genes, A leader sequence, A repeat spacer array.
What is Cas 9 and what does it stand for?
Cas 9 means crispr associated protein 9. It is the enzyme that uses CRISPR sequences to recognise and cut complementary DNA to these CRISPR sequences.
How was CRISPR-cas discovered
In 1987 in Japan, Yoshizumi Ishino identified repeats while studying genes from E.coli. In 1993 in holland, Gorenen discovered the diversity of intervening sequences while studying tuberculosis. In 1993 in Spain, Mujica identified repeats of Haloferax and Haloarcula. IN 2002 in holland, Hansen identified was genes which are associated with CRISPR repeats.
How does crispr cas 9 work simple
It works like small molecular scissors that can cut DNA at specific spots.
A guide RNA is created by scientists which matches the DNA which they want to change.
Cas ( enzyme. This Guide RNA is attached to Cas 9 enzyme which acts as the scissors which cut the DNA.
The guide RNA leads Cas 9 to the exact spot in the DNA.
Cas 9 cuts the DNA at that spot
After the cut, the cell tries to repair the DNA. Scientists use this moment to turn off a gene, fix a broken gene or to insert a new one.
What is PAM.
It is Protospacer Adjacent Motifs. They are very short DNA sequences that Cas9 must bind to. CAS 9 will not successfully bind to or cleave the target DNA sequence if it is not followed by the PAM sequence. It essentially allows CAS9 to differentiate viral DNA from its own genomic DNA.
List some of the different applications of CRISPR cas 9
Gene knock-out (switch off a gene).
Gene editing (modify a gene).
Transient gene silencing or transcriptional repression (CRISPRi).
Transient activation of endogenous genes (CRISPRa or CRISPRon).
Embryonic stem cell and transgenic animals.
Pooled genome-scale knockout screening.
Modification of organisms such as Yeast: biofuels, Plants: Improve crops, Animals: Mosquitos “anti-malaria”.
Re-evaluations related to gene-disease claims have been carried out.
What is Vector DNA and why is it required for DNA cloning.
Vector DNA is a DNA molecule which acts as a molecular carrier that brings the DNA of interest into the host cell and facilitates replication. This vector DNA is usually a plasmid.
Define the following terms: Artificial chromosome, DNA libraries, F factor and multiple cloning site.
Artificial Chromosome: is a functional chromosome created by genetic engineering, with a centromere and a telomere and it is transmissible in cell division.
DNA Libraries: a collection of fragmented DNA that is stored and propagated in a population of micro-organisms via molecular cloning.
F Factor: the fertility factor in bacteria, also called episome and involved in bacterial conjugation.
Multiple cloning site: Short sequence of DNA with restriction sites, usually unique in a vector.
What are the different types of vectors and how much is the difference in base length.
Some other ones are:
What are the 2 main varieties of Artificial chromosomes?
Yeast artificial chromosome and bacterial artificial chromosome.
Provide a definition for plasmids
They are small circular dsDNA that autonomously replicate independently of the chromosome of the host bacterial cell. They can be resistant to some antibiotics and they have an origin of replication which is a region of DNA that allows multiplication of the plasmid within the host.
What are bacteriophages. How long is their dsDNA sequence. What can they be used for.
They are a type of virus which are capable of infecting bacteria. They have a 50kb dsDNA sequence. They can be used to clone larger segments of DNA.
What is recombination based technology.
It’s a method to transfer DNA between plasmids using site-specific recombination instead of restriction enzymes and ligases.
What are the key elements used in Gateway cloning?
att sites (attB, attP, attL, attR) and recombinase enzymes (BP Clonase and LR Clonase).
What are the requirements for DNA cloning.
Foreign DNA (e.g. PCR, genomic DNA cDNA)
Host organism.
Vector DNA for cloning.
Means of inserting foreign DNA into the vector.
Method of placing the in vitro modified DNA into the host cell.
Methods for selecting and/or screening cells that carry the inserted foreign DNA.
What is T-A cloning directly from PCR product.
It relies on the ability of Adenine and Thymine on different DNA fragments to base-pair. A ligase molecule binds them together.
Why would we wish to clone.
In order to isolate DNA which can be sequenced. To establish collections of DNA libraries. To perform further molecular studies such as the production of proteins, investigating proteins, identifications of mutations e.g. gene defects related to specific diseases.
What is complementary DNA (cDNA)
It is DNA synthesised from an mRNA by reverse transcriptase and DNA polymerase. It is a clean open reading frame.
What is DNA cloning.
It is a technique where foreign DNA is inserted into a vector which is capable of replicating separately in a host cell usually e.coli. Growing the host cell allows the production of multiple copies of identical inserted DNA for use in a variety of purposes.
What are the requirements of ligation reactions.
2 or more fragments of DNA.
A buffer containing ATP.
A DNA ligase enzyme.
what is are some methods of transferring DNA into a host cell.
Electroporation: The membranes of the cells are permeabilised using an electric field. The DNA present is able to penetrate the membranes after the electric shock.
Conjugation: It is the natural transmission from donor to recipient. It involves the formation of cell to cell junctions.
Transfection of the DNA:
The DNA is packaged in vitro into phage particles. The phages are allowed to infect bacterial cells. The DNA in transiently expressed.
In what conditions are hosts grown in for the identification of positive clones.
Antibiotics, Nutrient requirements, plaque type, blue-white screening.
What is blue white screening.
Blue-white screening is a method used in DNA cloning to identify bacteria with recombinant plasmids. The plasmid contains a lacZ gene with a cloning site inside it. If foreign DNA is inserted, β-galactosidase is disrupted:
White colonies: contain the insert (recombinant)
Blue colonies: no insert (non-recombinant)
What does gateway technology use which is different to other methods of DNA cloning.
The Gateway® technology, is based on the bacteriophage recombination system. It uses a BP clonase and an LR clonase, not restriction enzymes and ligases.
What is DNA sequencing
It is the process of determining the nucleic acid sequence - the order to nucleotides in DNA. DNA contains the genetic instructions that all organisms use to function. These bases are Adenine, Thymine, Guanine and Cytosine.
Why is DNA sequencing important.
It helps evolutionary biologists to understand how organisms or genes are related to one another
It helps biologists to study individual genes and proteins by verifying the accuracy of molecular cloning experiments.
We can identify mutations behind certain diseases.
We can diagnose genetic diseases by sequencing patients DNA.
Diagnose infections by detecting or sequencing bacteria/viral DNA.
in genealogy uses such as 23 and me.
What is first generation sequencing techniques. What were some techniques used.
They are methods which were developed in the late 1970s. This can be madam Gilbert sequencing where up to 400 bases can be sequenced. Sanger sequencing can sequence up to 1000.
When did the human genome project generate its first sequence using Sanger sequencing.
2003.
What is second generation sequencing and what are some techniques which were used.
It is a method which sequences millions to billions of DNA sequences simultaneously. These techniques are also known as next generation sequencing because they sequence many fragments at once. Biologists use these methods to generate short reads into a larger DNA sequence or compare reads to a known genome. examples can be 454 pyrosequencing. It generates reads from 50-500 bases.
What is 3rd generation sequencing.
It was developed in the 2010s. It is also known as long read sequencing. It can provide reads od up to 10000 reads. They also use Sanger and Gilbert sequencing similar to first generation.
What does Sanger sequencing require
It requires a DNA primer, DNA polymerase, dNTPs (deoxynucleotide triphosphate), ddNTPS (dideoxynucleotide triphosphate) help with chain termination and the template DNA.
What are the steps of Sanger sequencing.
Denaturation and annealing.
It begins by separating the double stranded DNA fragments into 2 single stranded DNA fragments. A primer binds to the ssDNA based on complementary pairing between the primer and DNA.Extension
A mixture of dNTPs and ddNTPs are added to DNA polymerase. If a dNTP is added, extension continues. If ddNTPs are added, the extension stops. This causes the length of the DNA fragments to be different. ddNTPs also have fluorescent markers which help in the visualisation step.
Separation: Amplified DNA fragments are then separated by size using capillary electrophoresis. In this process, smaller fragments travel faster than larger ones.
Visualisation: The sequence is determined by visualising the fluorescent tag incorporated at each position of the DNA sequence. AS the DNA moves past detector, the laser emits light and the camera records the emitted light. The different levels of fluorescent lights produce different signals. This helps determine the different amount of bases in DNA sample.
What are some pros and cons of Sanger sequencing.
Pros
Simpler data analysis compared to NGS
Affordable for a small number of samples
Highly accurate
Longer reads compared to NGS (up to ~1 kb) so it can help improve accuracy and assembly of repetitive regions.
Cons
Requires a larger amount of input DNA
Low throughput
Unaffordable or impractical for sequencing a large number of samples
Why can next generation sequencing be preffered over Sanger in some cases.
It produces a larger number of DNA fragments. It is also more suitable for DNA samples with a low amount of starting quantity such as environmental samples.
What are the steps of NGS DNA sequencing.
DNA extraction: NGS isolates teh DNA using a kit. The kit type depends not eh sample source e.g. bacteria, tissue, blood)
Library preparation: DNA forms fragments. After fragmentation, adapter sequences are added to the ends of the DNA fragments. These sequences immobilise DNA fragments to a flow cell and contain sequences for sequencing primer attachment. This forms DNA library.
(3.1) Bridge amplification: DNA library is denatured and attached to the flow cell via adapter sequences. Flow cells have billions of spots which allow simultaneous sequencing of billions of fragments. These immobilised DNA sequences are amplified via repeated PCR cycles to generate thousands of copies of each original fragments. These dense clusters of identical sequence create a strong fluorescent signal that can be detected.
(3.2) Sequencing by synthesis. A sequencing primer which is complementary to the adapter sequence is required. DNA polymerase and chain terminating bases that contain dye. Each 4 bases contain different labels. DNA polymerase adds one base at a time. The dye is removed and next base is added. The fluourescence is recorded and it dermises the sequences for millions of different DNA strands in parallel.Sequence analysis: After sequencing, the DNA sequence is analysed using computational tools. Sequences are arranged by overlapping regions. If a reference genome exists, it can be used as a guide to align and map short reads to their correct locations.
What are the pros and cons of NGS
Pros
High accuracy compared to long-read sequencing (~0.01% error rate)
Lower cost compared to long-read sequencing
Cons
Unequal error rate across the read
Only capable of producing short sequencing reads (reads are between 200-300 bases long)
Difficult to assemble repetitive regions (may be larger than the NGS read length)
Requires large data storage capabilities and computational resources
What is long read sequencing. What are some disadvantages to this technique.
It generates long reads generally 10kb to over 50kb. These methods sequence a single DNA molecule without amplification steps. It typically has a higher rate of errors than short term sequencing.
What is nano pore sequencing. What are the steps involved.
It is a type of long read sequencing. It relies on the fact that each nucleotide has a different size and electrical property.
motor protein unwinds the DNA allowing it to pass through a pore on the membrane.
When a base passes through the pore, it reduces the ionic current.
Each base reduces the ionic current by a different amount. This means that the sequence of the DNA strand can be determined.
What are the pros and cons of long read sequencing.
Pros
Easier library preparation
Very long reads (up to 2Mbp on ONT)
Assembly becomes less ambiguous compared to assembling short reads
Portability. The platforms are about the size of a USB.
Fast sequencing runs
Epigenetic markers are stable and so methylation signatures and histone modifications are preserved
Cons
Higher error rate than NGS (ONT)
A common approach to overcome these errors is to attain a high level of coverage by sequencing the same site multiple times over, providing a consensus for each site.
In most cases, coverage of 30x–50x (each base is read 30–50 times on average) is sufficient for high-quality sequencing.
However, higher coverage comes at a higher financial cost.
When should these techniques be used specifically.
Sanger
ideal for small-scale projects focusing on one or two genes
Illlumina
high-throughput and DNA quality is not important (highly fragment)
PacBio
Costly but more accurate than ONT
Good for generating reference genomes
ONT
On the go sequencing
Population scale
Methylation
Define viruses. What features do they have genetically.
They are sub microscopic intracellular parasites ranging in size from 0.02 to 0.3 mm. The largest bacteria is smaller than the smallest bacteria. Viruses are biologically inert, they do not grow or undergo division. They lack genetic info necessary for the generation of metabolic energy.
When were viruses first thought to have has a written record.
Ancient Egypt, roughly 1400 bc. Rabies has been described and documented for more than 2000 years.
Provide some examples of plant, animal, fish, bacteria and insect viruses.
plant viruses: Tomato Spotted Wilt Virus
animal viruses: Foot and Mouth Disease Virus
fish viruses: Fish Infectious Hematopoietic Virus
Bacteria-T4 Bacteriophage
Insects:Baculovirus
What are the 2 main varieties of viruses.
Non enveloped (naked) and enveloped
Describe some features non enveloped virus particles often have
Capsid surrounds nucleic acid genome.
Self assembly of capsid from capsomeres
Have symmetrical shapes
Spheres
Rods
Often other enzymes are included,
Lysin (phage)
Why do non enveloped viruses have a caspid
They have a caspid to protect against physical, chemical and enzymatic damage. They also help the viruses to bind to receptors using spikes on their structure. They help transmit signals.
How do non enveloped viruses affect food.
Non-enveloped viruses can contaminate food and cause illness. They are tough and resistant to heat, drying, and chemicals, making them hard to kill during food processing. Common examples include norovirus and hep a. They can be found in livestock which caused the culling of 6.5 million cattle sheep and pigs. The food can be used as a vector to transmit viruses from animals to humans.
Describe some of the features an enveloped virus has.
Have an extra shell of lipid bilayer from the host, allows fusion with host’s cell wall.
Can exit the cell WITHOUT total destruction of the host
What are some consequences of viral infection of animal cells.
What is lytic infection
Lytic (acute) infections
host cells is destroyed-
virus particles are released & normally cleared within a couple of days, hosts develop immunity.
Effective strategy, some hosts are not immune.
Can cause epidemics, polio, Flu, measles especially in DORMATORIES, nursing homes, schools and offices
What are latent infections
Last the lifetime of the host.
a latent state is characterized by a lack of gene expression, normally resides in sensory and autonomic ganglia which is immunocompromised.
Reactivation, which can be caused by UV light causes reentry into lytic cycle.
What is the difference between genetics and genomics and gene editing.
Genetics: The study of genes heredity and the variation of inherited traits. It explores how inherited traits are passed down from parents to offspring.
Genomics: The study of an organisms complete set of genes and their interactions.
Gene editing: Technologies that allow for precise modifications to an organisms DNA sequence. It can be used to alter specific genes or to insert new ones.
How can genetic testing, modification and editing be used in animal health, food production and livestock production.
Genetic testing can identify disease susceptibility. We can enable preventative measures and improve breeding procedures.
Food production: Genetically modified crops can enhance yield, nutritional value and pest resistance. This contributes to food security.
Livestock production: selective breeding for desirable traits such as milk yield, meat quality and disease resistance can improve efficiency and profitability.
What are some ways variation can be observed in animals. What techniques are used.
Variation can be generated by genetics.
Selective breeding methods where organisms are bred with desired traits to produce offspring with those traits.
Artificial insemination using semen from an animal with the desired traits.
What factors affect milk production and composition.
Genetics/breed: Geen expression of milk fat and the casein gene.
Diet characteristics: Different food sources affect the yield. Forage/fibre/type/protein concentration.
Health/physiology
Individual animal factors. (age, state of lactation)
What are the 2 main milk proteins. What percentage do each of them make up. What are they responsible for
Milk contains Two Main Proteins
Casein
80% of milk protein
Micelles: cluster of molecules found in colloidal dispersions
Make milk white
Curds-casein clumps that separate from the liquid
Acid precipitable
Whey
20% milk protein
Protein found in liquid that remains after fat and casein have been removed from milk
Acid soluble
Which animal has the highest mean protein content per litre of milk
Rats which have 11.8mg/L
What are the different components of milk
Protein, Lactose, Minerals and vitamins. Milk is mostly water and the main solids are fat, protein and carbohydrate. 3.9% fat, 3.4% protein and 5.2% lactose and minerals.
What is lactose and why is it important.
It is the most common carbohydrate found in milk. It is virtually only found in milk. It is a disaccharide of glucose and galactose with a 1,4 glycosidic bond. It is a white solid which is fully soluble in milk. It can flip from alpha to beta form in solution. Different versions of lactose are used for different products.
Out of alpha and beta lactose which is more soluble.
Beta lactose is more soluble. The ratio of alpha to beta lactose in milk powder determines its solubility.
What are lactose, maltose, glucose and fructose all examples of
They are all examples of non-reducing sugars.
How many varieties of fatty acids are found in cows milk. How do they differ.
There are up to 200 types of fatty acids in cow milk. The order of length in their chains is how they differ. Shorter fatty acids chains can release energy more quicker than longer chain fatty acids.
Where do milk fatty acids often originate from.
They originate from diet, rumen and the desaturation of fatty acid enzymes.
What are teh functions of rumen, reticulum, omasum, abomasum, oesophagus and intestine.
1. Rumen – The largest compartment where fermentation occurs. Microbes break down fibrous plant material into digestible nutrients.
2. Reticulum – Works with the rumen to trap foreign materials and assist in regurgitation for “chewing the cud.”
3. Omasum – Absorbs water and nutrients from the partially digested food.
4. Abomasum – The “true stomach” where enzymes and acids digest food before it moves to the intestines.
5. Esophagus – Transfers food from the mouth to the stomach and allows regurgitation for re-chewing.
6. Intestine – Absorbs nutrients and expels waste.
What type of sugars is casein mostly made of.
its mostly made of short lengths of alpha helix and beta pleated sheet.
How do casein proteins form casein micelles.
They associate with calcium phosphate molecules. This causes them to cluster together as the calcium phosphate acts as a bridge which holds the proteins together in a spherical structure.
Wheat is lactogenesis, where does it occur. How can milk be removed.
Lactogenesis occurs at the lactating mammary gland. It’s the only gland which can synthesise lactose. Milk flows into the teat canal. It can be removed by suckling or milking.
Where do ruminants and non ruminants get their energy supply from.
Non-ruminants get energy from glucose via dietary absorption and gluconeogenesis.
Ruminants get energy from glucose and acetate.
Key stages of mammary gland development?
Mammogenesis, lactogenesis (1 & 2), galactopoiesis, involution.
Unique feature of lactating mammary glands?
Only organ that synthesises lactose
Major precursors for milk?
Glucose, acetate, β-hydroxybutyrate, amino acids, triglycerides.
Key microbial players in the rumen?
Anaerobic bacteria, fungi, protozoa, archaea.
What improves milk fat?
Increased acetate.
What does milk production rely on?
Hormonal signals & nutrient supply.
What does the equation P = G + E represent?
Phenotype = Genotype + Environment.
What are additive and non-additive variations?
Components of genotypic variation affecting traits.
Example of polygenic traits?
Milk yield, longevity, milk fat yield.
What is transgenesis?
Direct manipulation of DNA to insert genes from another source.
Benefits of transgenesis over selective breeding?
More efficient and precise.