4L4-6 Nitrogen Catabolism, Reactions, and Excretion
1/114
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
115 Terms
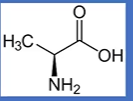
alanine
-CH3;
A = first in alphabet of the As;
np, Hphobic, aliphatic
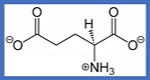
glutamate
-CH2CH2CO(O-);
E= gluEtamate ,Glu;
negative charged second carbonyl
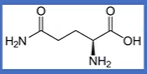
glutamine
-CH2CH2C(=O)NH2;
Q = Qtamine, amide of glutamate
Gln = Glu but with an n;
polar, uncharged, Hphilic
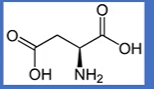
aspartate
-CH3CO(O-);
D = asparDate, you ASk for a Date;
negative charged second carbonyl
pyruvate
structure
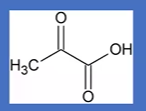
aKG
structure
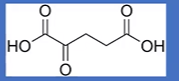
oxoaloactetate
structure
amino acid sources
intracellular proteolysis
dietary proteins
de novo synthesis
intracellular proteolysis
removes misfolded and damaged proteins
supplies essential amino acids when diet insufficient
de novo synthesis
nonessential for protein synthesis
amino acid pools in tissues
energy metabolism by controlling metabolites for central pathways
needed for NuTides, hemes, hormones, NeuroTs
essential amino acids
valine, leucine, isoleucine, tryptophan, phenylalanine, lysine, histidine, methionine, threonine
***MILK TV FHW
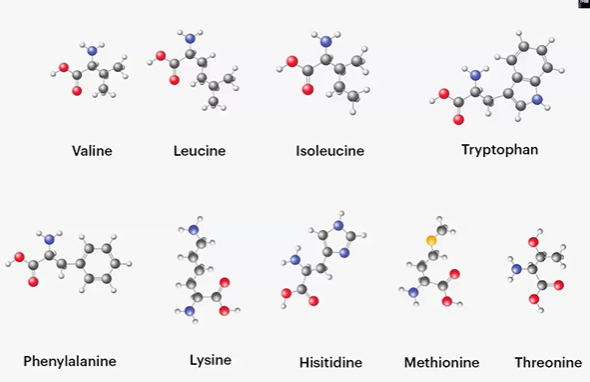
valine
-CH(CH3)2;
np, Hphobic, aliphatic
leucine
CH2CH(CH3)2;
L = alphabetically first of the Ls
leuc = 4C;
np, Hphobic, aliphatic
isoleucine
CH(CH3)(CH2CH3);
I = only I
leuc = 4C, iso = split;
np, Hphobic, aliphatic
tryptophan
-CH2 cyclo =C-N-C=Ph;
W = twyptophan, W before Y in alphabet,
polar, uncharged, Hphobic
phenylalanine
-CH2Ph;
F = fenyl;
polar, uncharged, Hphobic
lysine
-4CH2 NH3+;
K = next to L, Leuc = lys = 4, 4CH2;
positive charge, Hphilic
histidine
-CH2(NHCH=NCH=C) loop;
positive charge, Hphilic, ionizable side chain with neutral pKa
methionine
-CH2CH2SCH3;
Me = CH3, Thio = S;
np, Hphobic, aliphatic
threonine
CH-(OH)(CH3);
T = alphabetically first T, THREE (CH3) O (OH);
polar, uncharged, Hphilic
conditionally essential amino acids
we make it but occasionally need more
arg cys gln gly pro tyr
R Q G P Y C (real quick go produce your conditionals)
aa catabolism overview
sources proteolysis, digestion, or de novo synthesis
amino and carbon are separated
carbon > aKA > CAC
NH3 > biosynthesis / Urea cycle
combine with the asp-arg-succ shunt of CAC
ketogenic v glucogenic
ketone bodies v glucose
IC protein turnover
constant flux of synthesis / degradation
regulated by proteostasis
three machines: lysosome, ubiquitin proteasome, autophagic pathway
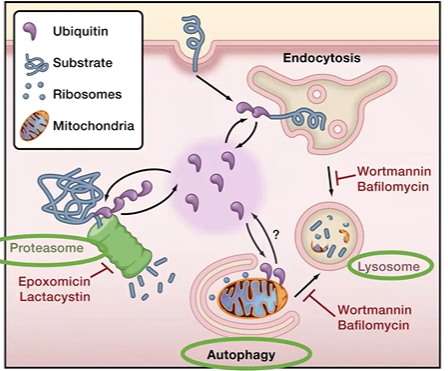
ubiquitination
adding ubiquitin to a protein to target it for proteolysis via proteosome
done by ubiquitin ligases, *1= Initiate, 2 = conJugate, 3 ligate
E1 initiates by grab Ub, pass to E2
E2 conjugates
E3 ligase, adds Ub tag
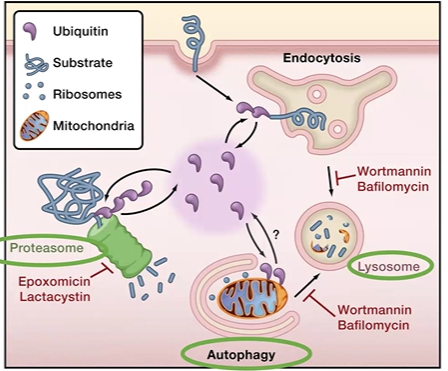
E1 E2 E3 ubiqiutin
1 = initiates,
2 = conJUgates
3 = ligases
***1 = i. two = ju
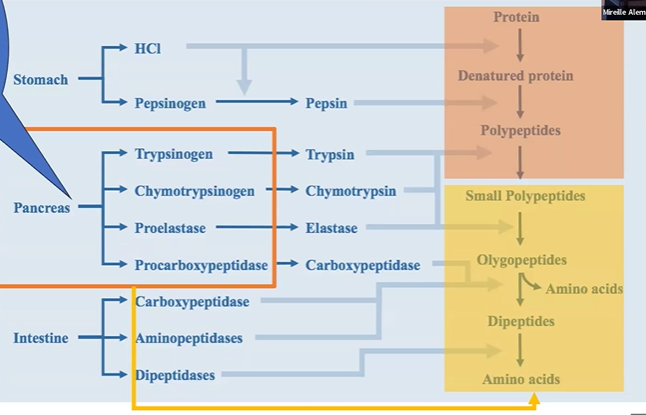
overall digestion process
first catalyzed by sol enzumes in stomach and SI
digestive enzymes in salivary, stomach, and pancreas
pancreatic enzymes and bile acids help intraluminal digestion
gastrin
secreted as dietary proteins enter stomach by gastric mucosa
stimulate HCl release by parietal cells
chief cells secrete pepsinogen
parietal cells
secrete HCl bc gastrin signalled
chief cells
secrete pepsinogen, inactive form of pepsin, low pH
autocatalytic cleavage
converts pepsinogen to pepsin for peptide breakdown
pepsin
cleaves polypeptides at low pH
cleaves phenyl alanine leucine or glutamic acid
secretin
secreted in low pH in intestine,
stimulates pancreas to secrete bicarbonate to neutralize HCl to 7
zymogens
inactive enzymes, cleaved to turn into active proteases
released by pancreas at pH 7
dietary protein degradation overview
chewing, amylase and lipase in saliva break carbs and lipids
proteins enter stomach, protonated by acidity,
gastric mucosa releases gastrin,
gastrin lowers pH with parietal cells’ HCl, also chief cells’ pepsinogen,
pepsinogen activated to pepsin at low pH,
pepsin cleaves aromatic polypeptide chains,
in intestine, secretin tells pancreas to release HCO3 to neutralize pH,
pancreas releases zymogens at ph 7, they turn into active proteases
trypsinogen + enteropeptidase > pi > alpha trypsin
chymotrypsinogen > chymotrypsin
procarboxypeptidase > carboxypeptidase
proelastase > elastase
pepsin activation
pepsinogen made and released in stomach
autocatalytic activation, acid catalyzed, low pH
helps activate itself to pepsin
activated by removing masking sequence
____is removed to turn pepsinogen into active pepsin
masking sequence
____ zymogen turns into _____ peptidase
trypsinogen > trypsin
chymotrypsinogen > chymotrypsin
proelastase > elastase
procarboxypeptide > carboxypeptidase
chymotrypsin cleaves at
carboxy side of aromatic residues
trypsin cleaves_____; activates ___
carboxy of lysine and arginine residues;
chymotrypsinogen to alpha
carboxypeptidase cleaves
one residue off carboxy end
proteolysis stomach overview
stomach makes HCl and pepsinogen
pepsinogen +HCl > pepsin;
protein + HCl > denatured protein
pepsin + denatured protein > polypeptides
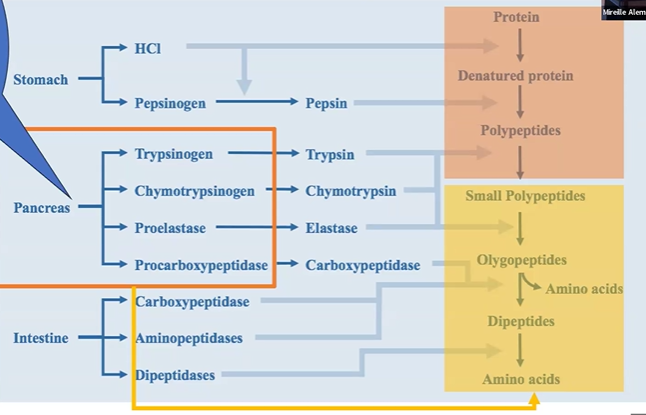
proteolysis pancreas overview
trypsinogen > trypsin
chymotrypsinogen > chymotrypsin
proelastase > elastase
procarboxypeptidase > carboxypeptidase
zymogen + polypeptide > smaller polypeptide / olygopeptides
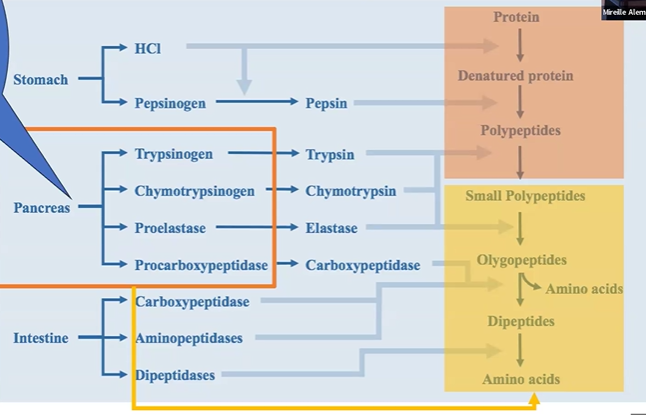
proteolysis intestine overview
carboxypeptidase and aminopeptidase turn olygopeptides > amino acids and dipeptides
dipeptidases turn dipeptides > amino acids
zymogens
inactive proteases made and stored in pancreas
secreted into small intestine via vesicles and converted to catalysts
_____inogen is transported in vesicles which also contain ____
trypsinogen, a trypsin inhibitor to prevent unwanted proteolysis
chymotrypsinogen
processed through sequential proteolytic cleavage
first to pi, second to alpha which is way more active
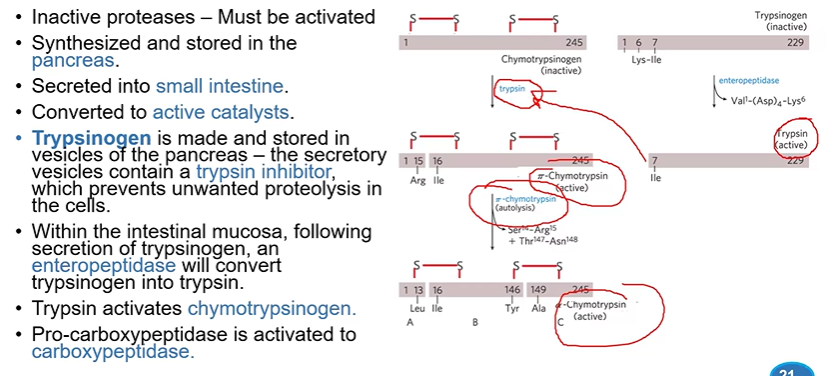
enteropeptidase
converts trypsinogen to trypsin
microvilli
increase SA and facilitate intestinal mucosa to absorb amino acids
form brush border cells
aa transporters
mediated by Na/K ATPase,
specificity for amino acids,
secondary active / facilitative transporters
from intestinal cells, enter blood, taken to liver
aa oxidation
deamination, not needed for synthesis or needed for energy (bad)
unless reused, amino groups channeled into single excretory end product (pee)
dietary requirements
0.8g protein per kg of body weight.
RDA = amount to achieve zero nitrogen balance
nitrogen balance
consumption = excretion, no net change
positive could be in children / pregnancy
negative is in starvation or trauma
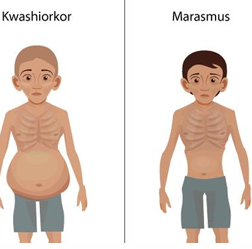
kwashiorkor / marasmus
childhood malnutrition, large belly, edema
lose skin, underweight, inadequate intake of proteins
essential amino acid
cannot be synthesized
proteasome
Large protein complex that degrades ubiquitinated proteins in a regulated way (selective degradation in cytosol/nucleus).
pepsinogen
Inactive zymogen secreted by the stomach that is converted to pepsin by low pH.
trypsinogen
Inactive zymogen from the pancreas; ****activated by enteropeptidase.
keto acid
Carbon skeleton of an amino acid after deamination—can be used for energy, gluconeogenesis, or fat synthesis.
most ingested proteins turn into
free amino acids in the liver
transamination
removal of nitrogen / amino group, deamination
almost always begins with transfer to aKG to yield glutamic acid
for all amino acids except for proline, lysine, and threonine
amino acids that don’t participate in transamination
proline, lysine, threonine
**Please, lets transmainate
aKG in transamination yields
glutamic acid
almost all amino acids use ___ to deaminate
aKG
amino acid + alpha keto acid =
new alpha keto acid + new amino acid
a-Amino group transfer uses enzyme ____ and ___, which carries ____
aminotransferase / transaminase, PLP, CO2
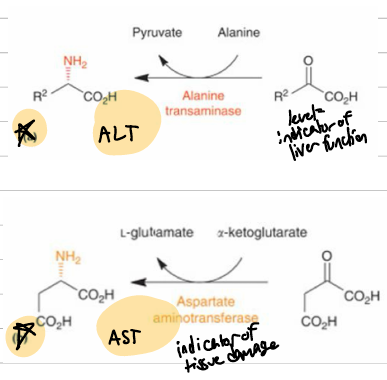
important aminotransferases / transaminases
AST = aspartate; aKg > L-glutamate; high in liver heart and muscle, indicator of muscle damage in high quantities
ALT = alanine > pyruvate; can release into blood in high concentrations. signifies excess liver usage, meaning damage
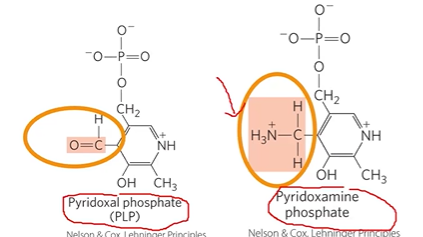
PLP
pyridoxal phosphate, prosthetic group
in all of pathways for amino degradation, protects release of amino group
PDP: C=O > NH3
Vitamin B6,
*looks like if a P and an L had a baby
PLP enzyme overview
AA1 + KA2 > KA1 + AA2
internal (enzyme bound) aldimine > external (amino acid bound) aldimine
DP and rearranges, quinonoid formation
rearrangement to help resonance
hydrolysis to aKA and PMP
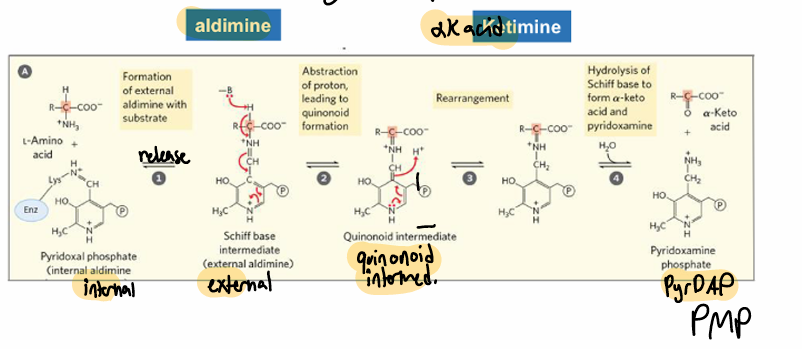
PLP starts ___ linked to residue on enzyme, this is an ____ aldimine
covalently;
internal
after amino acid approaches enzyme, PLP forms an ___ because amino acid ____s it
external aldimine;
activates
external aldimine becomes a ____ because of ____
quinonoid intermediate;
deprotonation
quininoid intermediate becomes ____ by ___
PDP + aKA;
rearrangement and hydrolysis
alanine aKG transaminase steps
alanine + PLP + H2O > PMP + pyruvate
PMP + aKG > G + PLP
*same as PLP but uses aKG specifically
aminotransferase reactions make what byproduct
aKA, often pyruvate
proline and hydroxyproline exceptions
secondary amines cannot react, only primary amino groups
lysine and threonine exceptions
lysine releases a toxic aKA metabolite (cyclic)
threonine releases a toxic aKA metabolite (dimerize)
glutamate dehydrogenase GDH
oxidative deamination; L-glutamate > aKG, helps renew ala-aKG transaminase
produces NADH and NH4+ ammonia
in mitoC matrix of liver
reversible, has intermediate
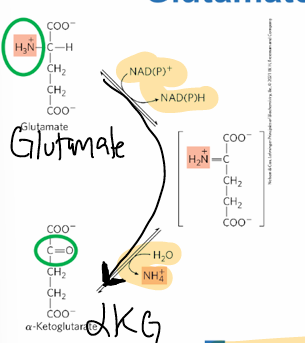
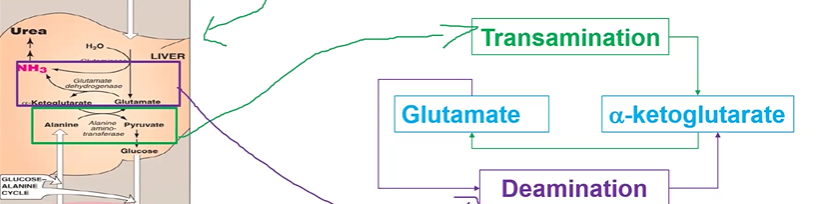
glutamate acts as a gate between
AAs and NH4+
coupling transamination and deamination
trans = aKG > glutamate
de = glutamate > aKG
in liver
GDH regulation
*high energy inhibits anabolic (protein synthesis, reduction), low energy activates deam (catabolic, oxidation)
inhibit protein synthesis = ATP, GTP, NADH
activate deamination = GDP, ADP, FAAs
glutaminase
deamination mitoC enzyme, glutamine > glutamate
other route for deamination
produces energy and ammonia
asparaginase
asparagine > aspartate + NH3
other route for deamination
amino acid oxidases
L and D, stereospecific, FMN or FAD involved as cofactor in redox rxn
H2O2 is a byproduct, which is highly regulatde
***other deamination
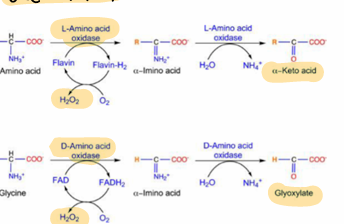
transdeamination involves ___ being transferred to ____ by ______, followed by release of ____ from ____ by _____
aA groups; glutamate; transaminations; NH4+; glutamate; L-GDH
ureotelic
pee out ammonia
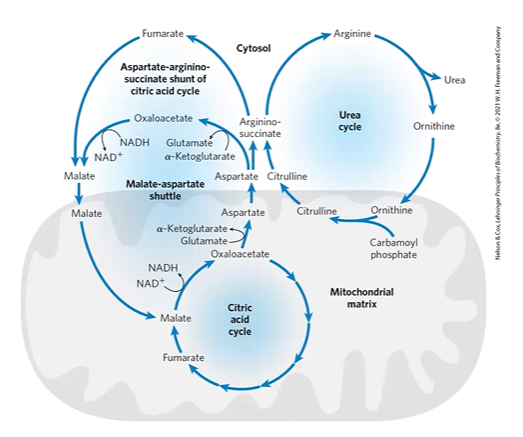
urea cycle begins in ___ but has steps in ___
liver mitoC; cytoS
glutamase GMAse
glutamase + glutamine from extrahepatic tissue > glutamate + NH4+
glutamate dehydrogenase GMDHase
GMDHase + Glutamate > NH4 + aKG
CPS1 CPSI
carbamoyl phosphate synthetase I
CPS1 + NH4+ + HCO3- + 2ATP > carbamoyl P
in mitoC activates urea cycle,
urea structure
NH2-C=O-NH2
carbamoyl phosphate (step 0 urea cycle)
enters urea cycle mostly in liver
activates ornithine trans carbomylase OTCM
NH4+ + CPS1 > CP
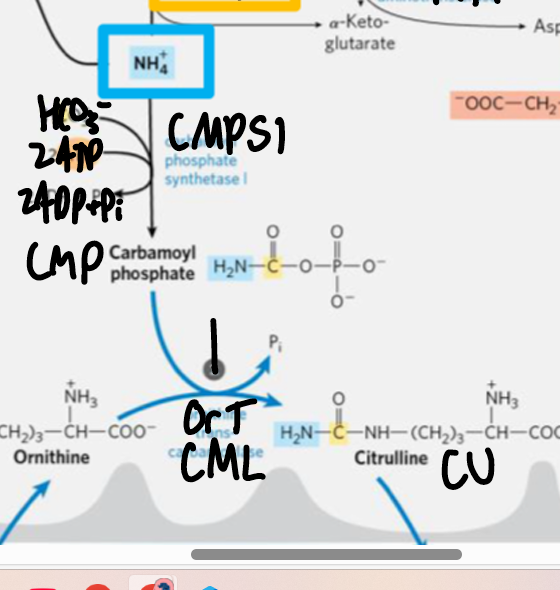
step 1 urea cycle
OCMP + ornithine + CMP > citrulline
**structures
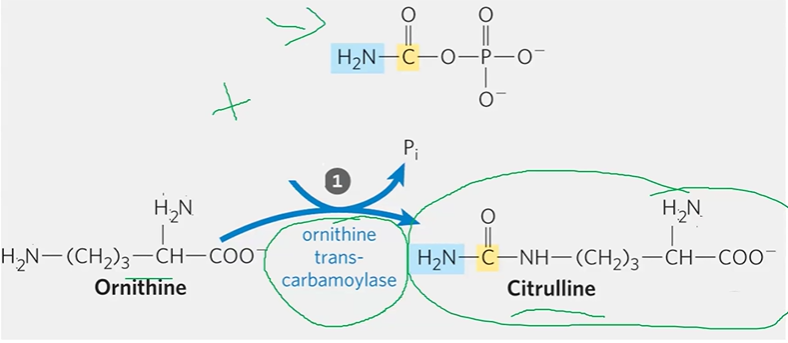
step 2 urea cycle
citrulline facilitated diffusion into cytoS
ArgSStase + Citu + ATP > int + Asp > ArgS + AMP
**structures
step 3 urea cycle
ArgSSase + ArgS > F (> malate > CAC) + Arg
step 4 urea cycle
Arginase + Arg + H2O > Ornithine
transported back via diffusion
urea cycle review
CO2 from CAC + ATP from OP + AA transported and aminated > glutamate,
glutamine > glutamate through GDH,
NH4+ + CO2 > CMP,
CMP + Ornithine + ATP > citrulline
citrulline + Asp > ArgS
ArgS > Arg + F
Arg > ornithine
*OCAsAO sounds like acacia
links between paths
urea cycle needs 3 ATP,
asp-arg shuttle of CAC works with urea cycle to make ArgS
regulation of urea cycle
at enzyme synthesis;
high protein diet; ala, gly, met, cys help glucagon, which stimulates biosynthesis
AA catabolism reduces NH4 load, increases OA and aKG for gluconeogenesis
glu and arg help synthesis of NAG
CPS1 is first committed step and highly regulated, allosteric activate N-Aglutamate
NAG n-acetyl glutamate
glutamate + NAGS + Arg > NAG + CoASH