Bio 2 Unit 3 Vesicular Transport
1/86
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
87 Terms
vesicular transport
moves material between the orange spaces (perinuclear space, ER, Golgi, endosomes & lysosomes, extracellular space) including liquid and membrane
Fuse and bud from plasma membrane → constant back and forth. material in the ER can be moved to other compartments, eventually being secreted into the extracellular space
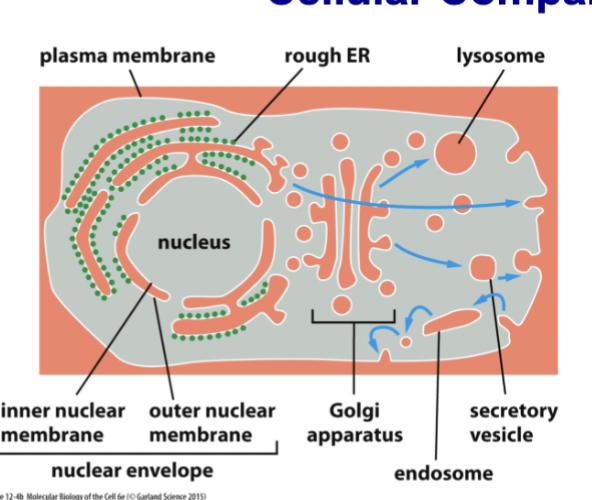
vesicles
small, spherical, liquid-filled, membrane-enclosed structures. They move proteins and other molecules between compartments
“ball of protein (inside + in membrane) held together by a bit of membrane (lipid bilayer)”
vesicle membrane proteins
cargo binding proteins, adaptor proteins, targeting proteins
coat proteins
clathrin, COPI, COPII
vesicle budding
membrane pinching off (collapse if it doesn’t complete in time)
vesicles are stable structures with a half life
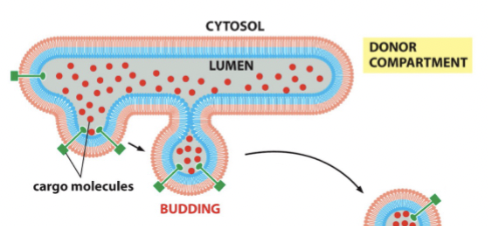
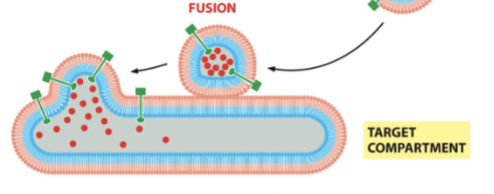
vesicle fusion
the vesicle gets very close to the membrane and initiates a _______ event where the inside of the vesicle is engorged into the target.
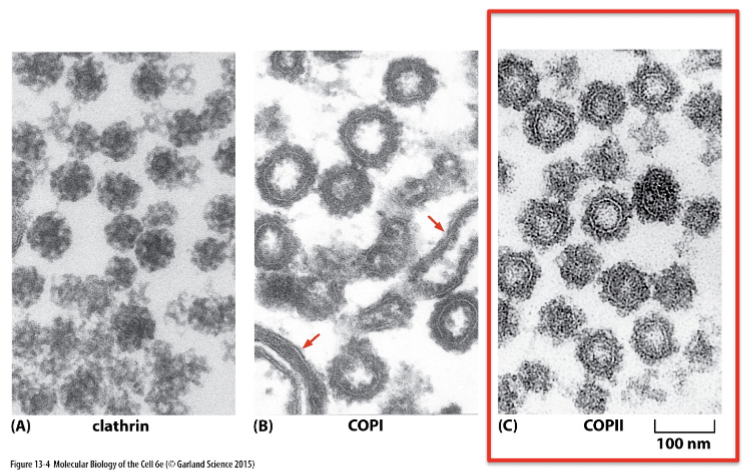
clathrin, COPI, COPII
three main coat proteins: each have a different distribution and mediate different transport steps
Each move different cargos, but the mechanism of budding and fusing is similar
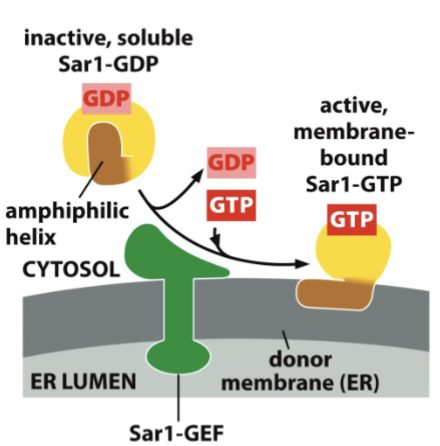
Sar1
insertion of this G-protein into the membrane nucleates vesicle assembly. Can move in and out of the membrane
the GTP form is membrane bound, and the GDP form is soluble. Regulated by corresponding GAPs and GEFs
Recruits other proteins to make the vesicle. Later, hydrolysis of GTP form to GDP form initiates coat disassembly
COPII vesicle formation
When Sar1 is bound to GDP, it has an amphiphilic helix folded inside the protein. Once bound to GTP, the helix pops out and is hydrophobic in places, allowing the helix to integrate into the membrane by inserting itself into the cytosolic leaflet of the lipid bilayer. The inner area remains the same, while the outer is larger, causing deformation of the membrane required to pinch off this vesicle.
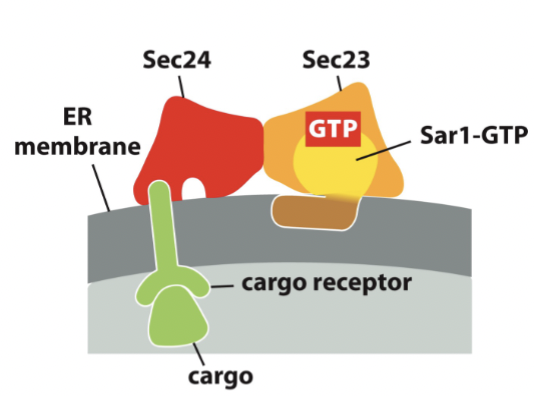
proteins Sar1-GTP recruits
inner coat proteins (Sec23 and Sec24), cargo proteins via cargo receptors, and other necessary protein → building large protein complex to be anchored in the membrane
delayed GAPs
coat proteins act as ___________ for Sar1-GTP: sets a timer to assemble the entire vesicle. If anything goes wrong, everything will fall apart before the vesicle can form, otherwise when Sar1 leaves
deform
assembly of coat proteins begins to ______ the membrane, causing it to look like a vesicle as more coat proteins are recruited
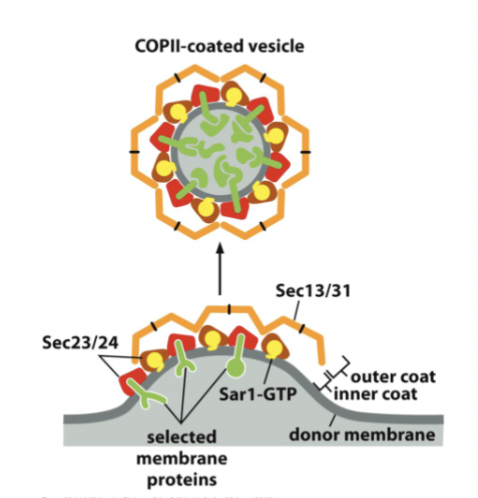
Sec13 and Sec31
after Sar1-GTP acts, these two outer coat proteins are recruited and bind to inner coat proteins, causing further deformation of the membrane (including cargo proteins)
regulatory molecules in COPII vesicle formation
v-SNARE proteins, membrane binding proteins, and membrane markers
bunch of proteins on and in the vesicles that need to be present during assembly before the vesicle can properly pinch off.
outer coat COPII
made up of Sec13/31, and binds to Sec23/24. Helps deform membrane. Can self-assemble into cages. Must be removed before vesicle can fuse.
Hydrolyze the GTP → Sar1 comes out of the membrane and lose Sec23+24, forcing these types of proteins to come off as well.
steps of COPII assembly and disassembly
Sar1-GTP inserts into membrane
binding of COPII inner coat proteins Sec23/24: requires presence of cargo proteins and Sar1-GTP and activates slow GTPase function of Sar1 à timer
outer coat proteins Sec13/31 bind, inducing more curvature
use GTPase function as a timer: at specified time interval, Sar1-GTP becomes Sar1-GDP, and Sar1-GDP causes Sec23/24 to fall off, outer coat falls off, Sar1-GDP leaves membrane
everything collapses, or the assembly occurs before the timer is up
vesicle budding then is a race between assembly and disassembly
coat must fall off of vesicles before fusion is possible
initiation and coat proteins
how does a vesicle form?
coat proteins act as coincidence detectors
how does a vesicle ensure the presence of cargo proteins?
SNARE proteins
control fusion, interaction between vesicle (single protein) and target (bundle of 3) proteins. They pull the vesicle close to the membrane by forming a helical bundle and use energy from the binding of these proteins. To unwind and recycle, they need ATP.
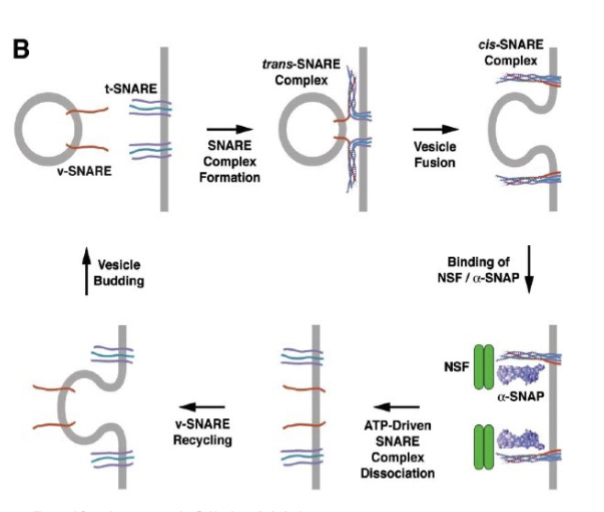
SNARE complex dissociation
driven by ATP, similar to charging a spring. the T-SNARE interacts with the V-SNARE, and the pent up energy is used to pull the membranes close together for fusion, and then the unbundling of the SNARES to be recycled is ATP-driven
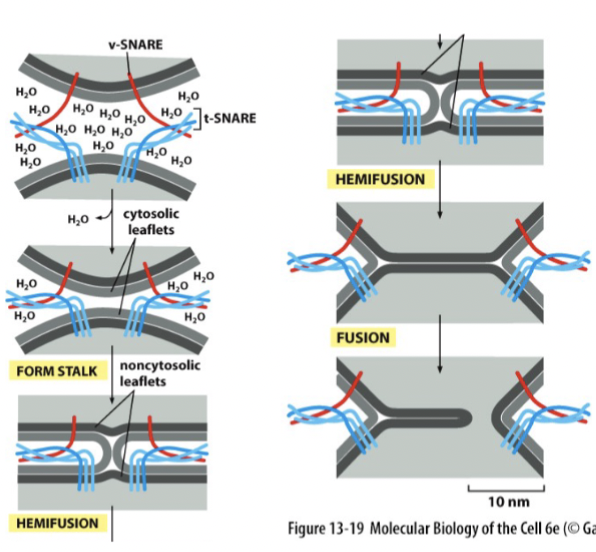
membrane fusion
SNARE proteins are like “pre-wound springs”. They coil together and bind tightly, and the released energy expels water molecules from the interface. If the membranes are 1.5 nm apart, lipids can flow from one membrane to another (regulated at synapse - happens automatically without any other protein regulation)
The unwinding step ‘recharges’ and recycles the SNAREs requires protein NSF and ATP
vesicle summary
small ‘balls’ of membrane; move cargo proteins between compartments, including soluble proteins and transmembrane proteins. Carry lots of proteins in membrane
think: many proteins held together by a little bit of membrane
budding requirements
coincidence detection (right coat proteins, cargo presence, …) and energy (membrane deflection)
fusion requirements
coat proteins removed, “spring-loaded” v- and t-SNARES, and energy required for SNARE recycling
v- and t-SNARES
how does a vesicle fuse?
coincidence detection
a contributor to specificity.
Only at the correct membrane patch, do you have sufficient amounts of all of the following (coincidence) to form a vesicle before the timer runs out: Coat Proteins, Cargo Proteins, Phosphoinositides (membrane markers), and Adaptor Proteins (required for some coat proteins
There is a limited number of places where everything is in place so things can form
microtubules
another contributor to vesicle specificity. Vesicles do not diffuse freely, they move along these.
not evenly distributed
coat proteins are … throughout the cell. Different coat proteins mediate different transport steps and are therefore located in different places in the cell.
phosphoinositides
make up about 10% of all phospholipids in the membrane. The inositol sugar (6-carbon, each has hydroxyl group) is heavily modified by phosphorylation from regulated (and specific!) kinases and phosphatases.
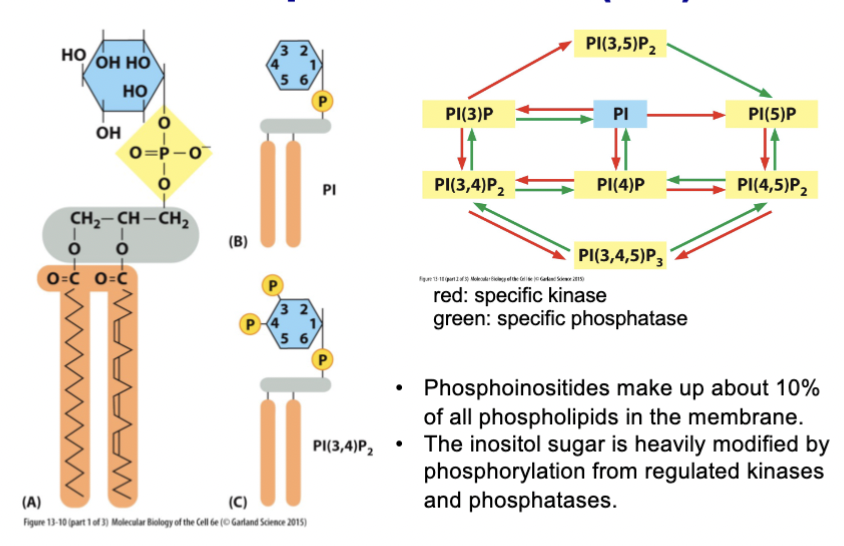
PI isoforms
binding proteins recognize specific _________, and there is regulated localization of these as well, and of their associated kinases and phosphatases. Vesicles budding in one place carry different _________ compared to other areas, and they create a code of membrane identity → budding specificity
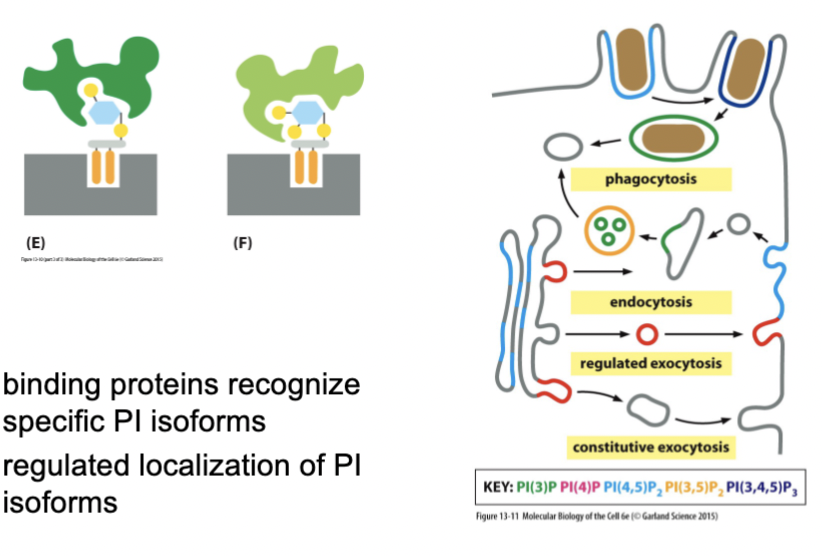
co-incidence detectors and phosphoinositides
how does a vesicle form in the right place?
The coat is required to bind to the membrane, and they only bind to specific PI isoforms when there are all the necessary proteins around
Rab proteins
contribute to fusion specificity
small G proteins, control activation of PI 3-kinase (controls phosphoinositides)
different family members → different subcellular localization in the family. Rab family proteins are present on target and on vesicle, but the specific Rab protein may be different on target and vesicle
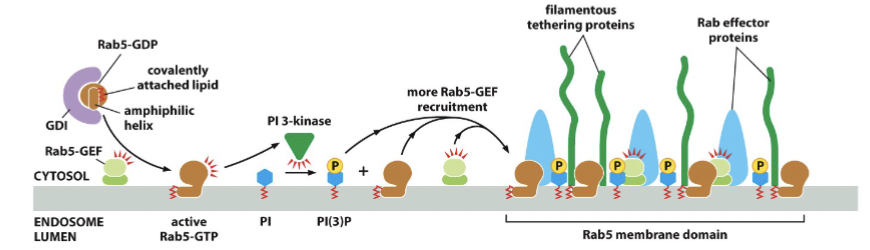
Rab5
Rab5-GEF activates this to become Rab5-GTP, which then binds to the membrane. Then the complex is disassembled by using Rab5-GAP to get Rab5-GDP
Rab effectors
there is a clustering of these on the membrane to give identity to that patch of membrane, including tethering proteins. Includes specific Rab-GAP protein → when a vesicle fuses with the target membrane, GAP makes sure the Rab protein exits the membrane, maintaining the identity after many fusions
Include: PIs, kinases, phosphatases for maintenance
tethering proteins
fishing rod proteins. Long, skinny, reach far into the cytosol. They want to find corresponding Rab-GTP in the vesicle and pull that vesicle in close enough for SNARE to interact for fusion.
These help initiate fusion by making sure vesicles come close enough to interact with the SNARE proteins
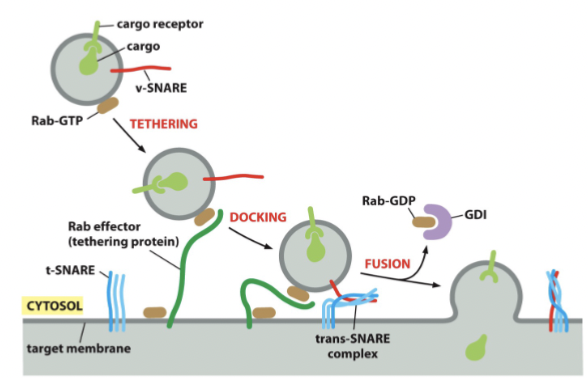
Rab proteins summary
cycle between membrane and cytosol
held inactive by Rab-GDP dissociation inhibitor (GDI) in cytoplasm
activated by special membrane-bound GEFs
activation causes Rab to bind to membrane
membrane-bound Rab activates Rab effectors, such as tethering proteins
tethering proteins contribute to fusion specificity
GDP dissociation inhibitor (GDI)
found in cytoplasm. functions in vesicular membrane transport to recycle Rab GTPases
solubilizes prenylated Rab GTPases from and shuttles them between membranes in the form of a soluble cytosolic complex, holding the Rab proteins inactive
SNARE and Rab proteins and phosphoinositides
three main pathways control fusion specificity; the distribution of each of these.
Together, pathways give identity to the origin of the vesicle, the destination of the vesicle, and the identity of the target membrane
redundancies
in the three pathways that control fusion specificity, there are multiple, built-in __________ so the cell can tolerate mis-match in any pathway and still get 100% specificity
distribution of SNAREs, Rabs, and phosphoinositides
how does a vesicle fuse with the right compartment?
vesicle formation and fusion
coat proteins assemble on membrane
recruit cargo proteins and regulatory proteins
vesicle buds, if formation faster than breakdown
coincidence detectors ensure:
site of budding
carrying of cargo
fusion
multiple, overlapping mechanisms ensure specificity
coat proteins, Rabs, SNAREs, phosphoinositides
additionally, vesicles are limited in their movement by being attached to microtubules.
pancreatic exocrine cells
produces many secreted enzymes for intestines, including pancreatic amylase, chymotrypsin, and digestive enzymes
George Palade
incubated slices of guinea pig pancreas cells with radioactive leucine
Pulse-Chase experiment
The pulse can be any readily traceable marker, such as radioactivity, creating bolus of material to follow through the cell. Today, fluorescent proteins are frequently used (some can be photoactivated + easily traceable)
Pulse with radioactive leucine (follow secreted enzymes, hormones, in pancreatic cells)
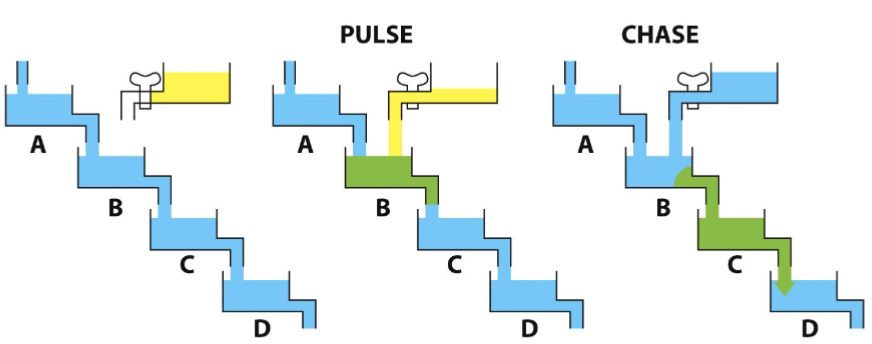
Palade’s experiment
radioactivity moves throughout the cell in a particular pathway.
“pulse” cells with radioactive leucine (leucine-3H) for 3 min
wash out radioactive leucine, replace with normal media containing unlabeled leucine
“chase” cells for 7, 17, and 117 min
fix cells
overlay with radiographic emulsion
analyze distribution of radioactivity → expose for about 2 weeks…
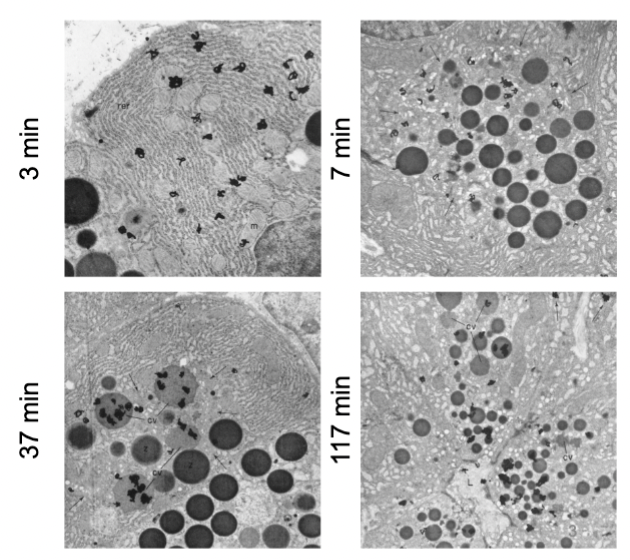
ER to Golgi to granules to extracellular space
Palade’s experiment established this direction of cellular flow .
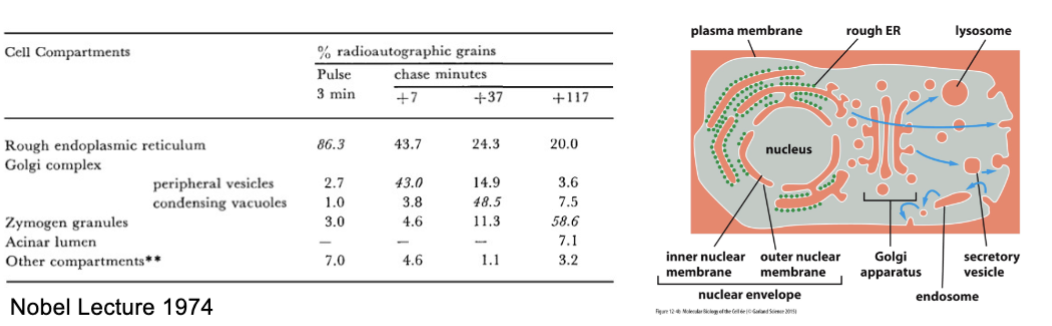
anterograde
ER to extracellular space (default)
extra signal
can put an _________ to prevent from going to EC space in anterograde . Sugars, proteins, have this to escape endosome in retrograde.
retrograde
endosome to ER. Safety mechanism: recycle unknown/unneeded material. The cell does not know what it takes up from the extracellular space and it does not want anything imported directly into the ER - so it destroys unidentified things in the endosomes.
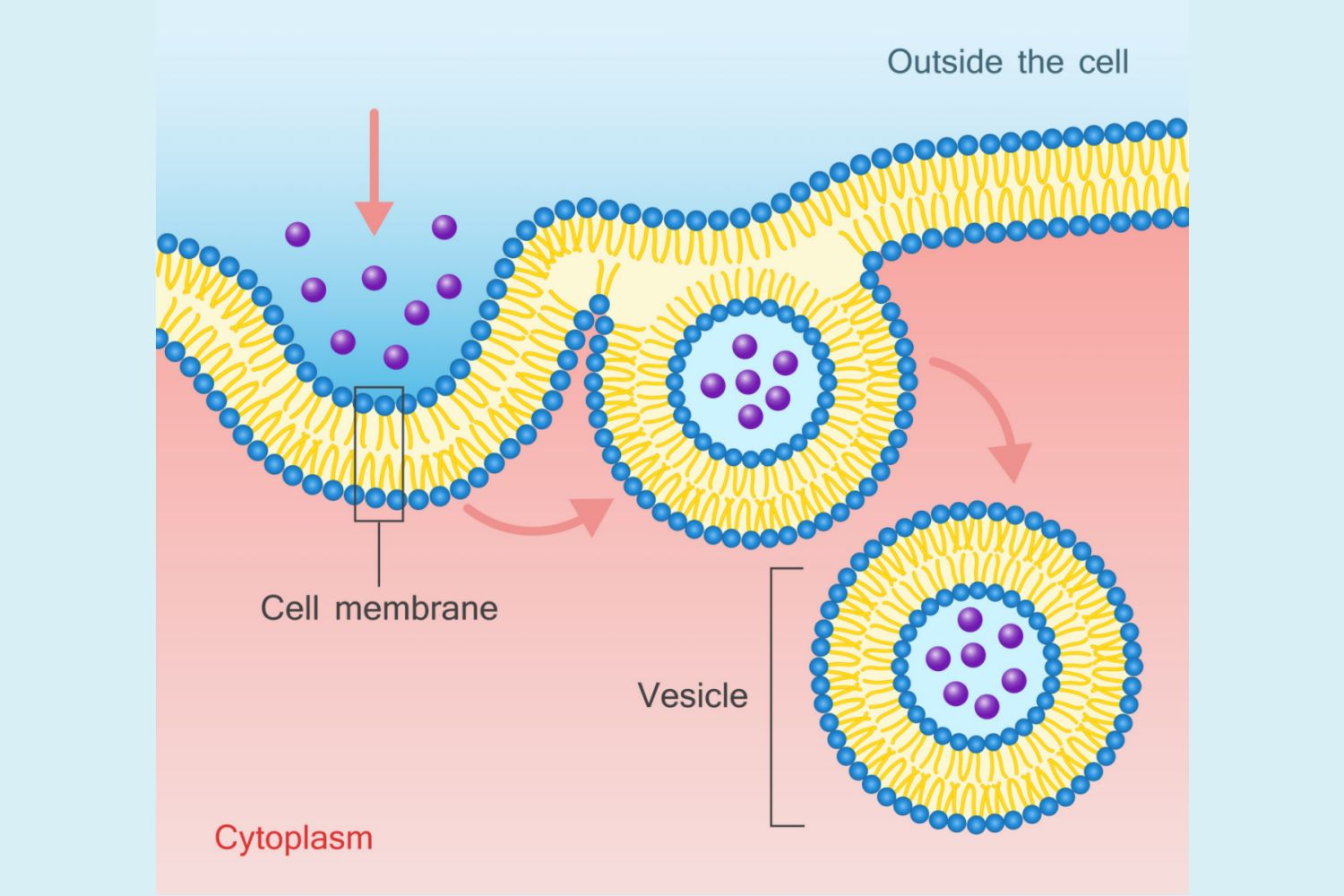
endocytosis
uptake of extracellular material.
a specialized form of transport by which very large molecules and insoluble materials are engulfed by invagination of the cell membrane forming intracellular vesicles
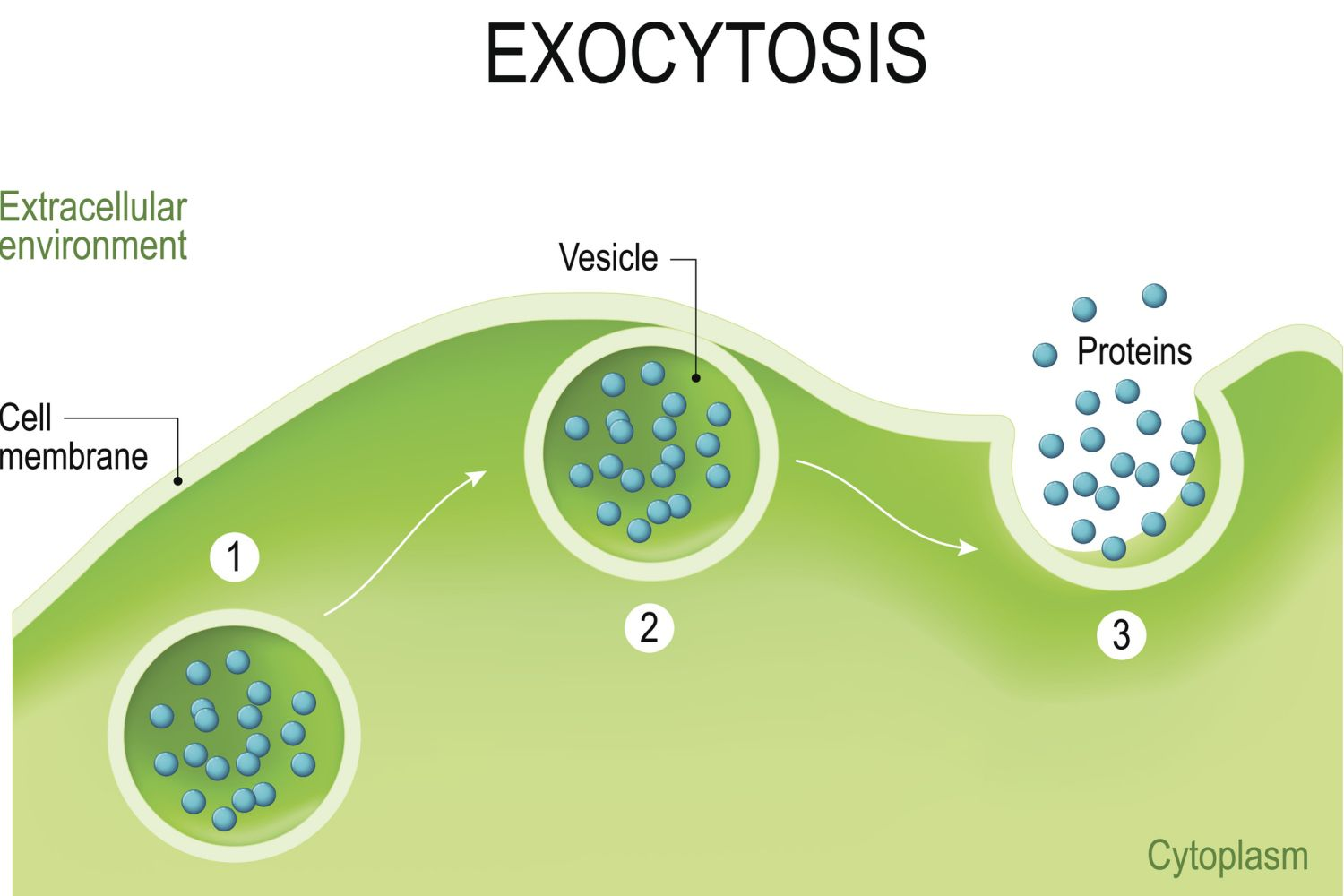
exocytosis
secretion into extracellular space.
a process for moving large molecules out of the cell to the cell exterior. Commonly, these macromolecules originate in storage vacuoles inside the cell and are moved to the exterior after an appropriate signal for this action
regulated transport
fast, needs receptors + signals
constitutive transport
slow. Everything just moves from the ER to the membrane without additional signals (go with the flow)
topologically identical
vesicles move proteins and liquid: the inside of the ER, Golgi, and vesicles are ___________ to the extracellular space
Liquid exchange → same internal fluid
exocytosis pathway
to extracellular space/plasma membrane. Any other destination needs extra signal
ER → Golgi → EC Space
endocytosis pathway
all material moved to endosomes/lysosomes. Diverting material needs extra signal
EC Space → Endosome → Lysosome
ER function
protein folding & processing and glycosylation (start)
Golgi function
glycosylation (finish), protein maturation, and sorting (materials are diverted to vesicles to specific places)
endosome function
material retrieval and sorting. Some material is re-exported, some to the Golgi or ER.
lysosome function
digest biological molecules. Anything without a signal is sent here for digestion.
folding and processing
Chaperones assist in protein folding. The formation of disulfide bonds by protein disulfide isomerase, the attachment of sugars (glycosylation), and the attachment of lipids to proteins are all parts of these functions of the ER.
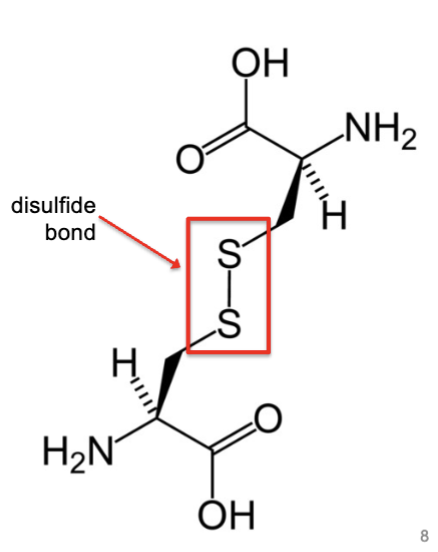
disulfide bonds
formed by protein disulfide isomerase, which lives in the ER.
formed by the oxidation of sulfhydryl groups between two cysteine side chains, resulting in a covalent bond, greatly increasing the stability of the protein
glycosylation
sugars transferred as a group (en bloc). Two forms: N-linked and O-linked. Means to hold the protein inside the ER.
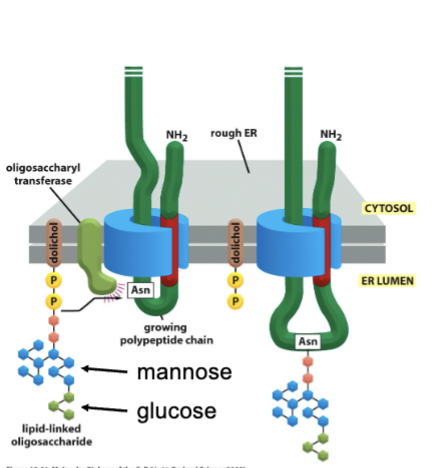
N-linked sugars
bind to N of asparagine, and occurs in ER. This type of glycosylation is required for many proteins to fold properly, but it can occur on any N of asparagine
O-linked sugars
bind to O of serine or threonin, and occurs in the Golgi
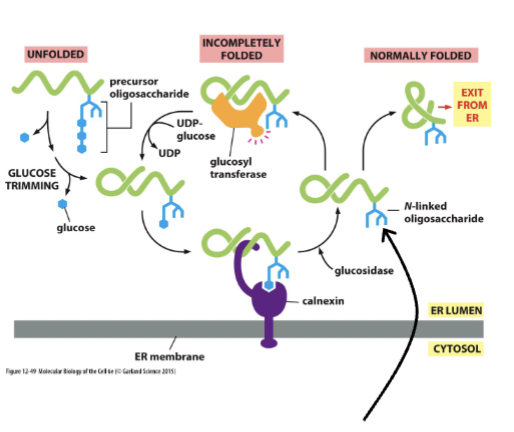
calnexin
glycosylation aids in the flor of liquid and biomaterial out of the ER to the Golgi, and glucose binds this compound, which works to retain unfolded/partially folded proteins in the ER to assist in folding (glucose removal and addition)
mannose
monitors total time spent in Er. Removal of this leads to NO addition of glucose. If the time is too long, the protein is exported for degredation
mannosidase
removes one of the mannoses after a certain time to target a protein for degredation
glycosylation and protein folding
this works to label proteins that can’t fold correctly → they are then pushed back into the cytosol and reduced to amino acids + recycled.
unfolded proteins in the ER
the proteasome degrades misfolded ER proteins. Stress response monitors how many misfolded proteins there are in the ER, and cancers are shown to be more aggressive if there is a higher concentration of …
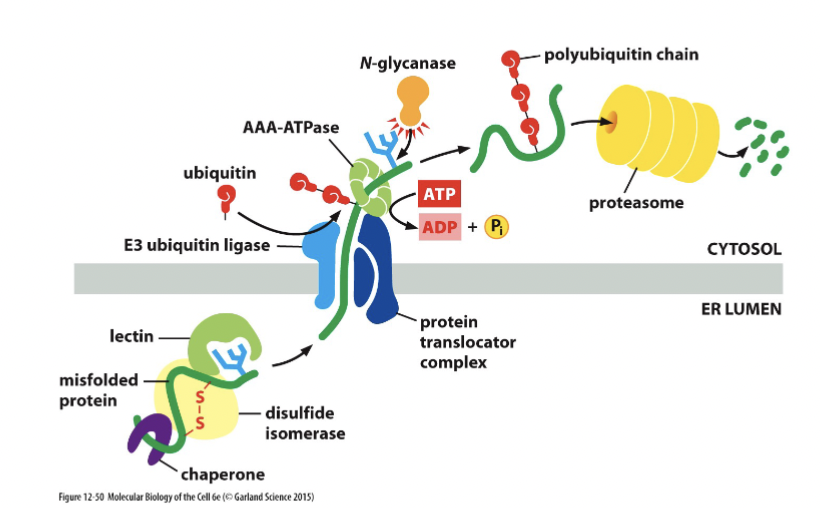
protein maturation in the ER
All proteins are glycosylated in the ER lumen by transfer of an oligosaccharide block to N of asparagine (any asparagine).
chaperones hold onto the sugars and assist in protein folding
incorrectly folded proteins are exported out of the lumen of the ER and destroyed by the proteasome in the cytosol
disulfide bridges form
ER stress
excessive levels of un- and misfolded proteins
correlates with worse prognosis for certain diseases
cellular response mechanisms to improve folding efficiency and removal of misfolded items.
Golgi stack
consist of a series of flattened curved and parallel series saccules, called cisternae or dictyosomes, that form the central portion of the Golgi complex. The stack usually comprises cis (faces ER + nucleus), medial, and trans (faces plasma membrane) cisternae
Has a particular direction/orientation. Directed flow inside - mediated by vesicular traffic.
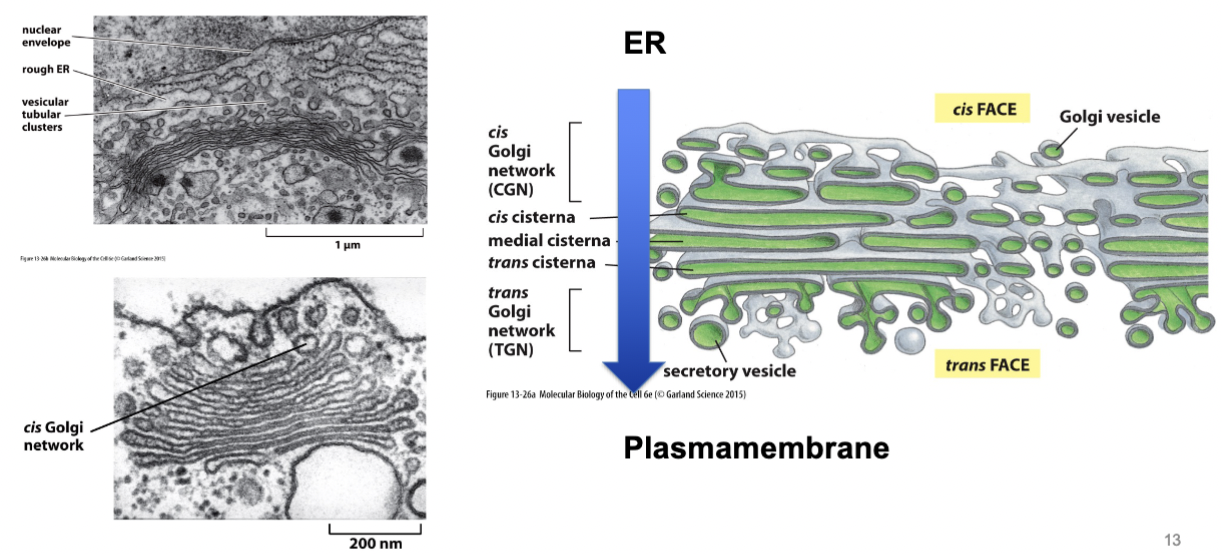
transport between ER and Golgi
smooth ER to cis Golgi, and both anterograde and retrograde transport.
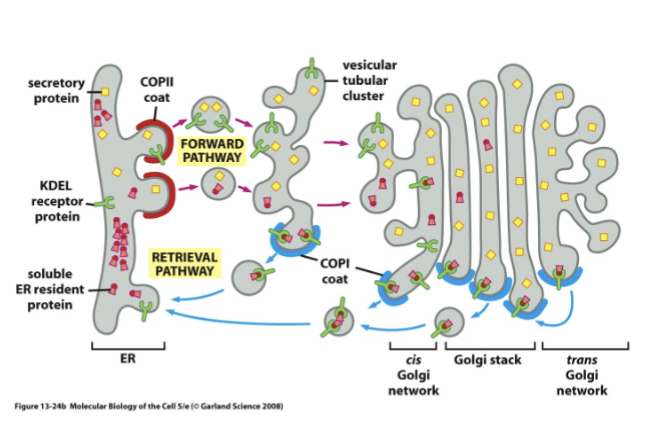
anterograde ER to Golgi
default secretion pathway (flow)
retrograde ER from Golgi
signal-mediated return of ER proteins that moved to the Golgi by the default transport pathway
some proteins escape the Er and are moved into the Golgi by accident and have a specific signal sequence that receptors recognize, and they are returned to the ER via this pathway
KDEL
this signal sequence indicates that something needs to come back to the ER (includes solbule and membrane proteins).
These sorts of receptors → seven-transmembrane-domain proteins primarily involved in the retrieval of ER-resident proteins from post-ER compartments to the ER lumen
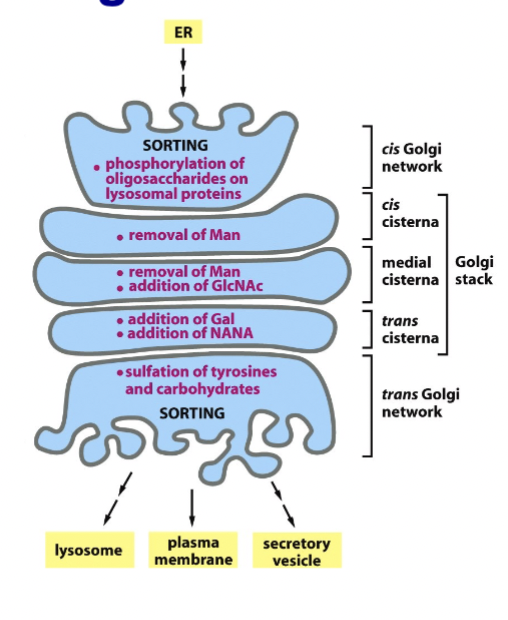
glycosylation in Golgi
N-linked glycosylation completed, and O-linked glycosylation started and completed
sulfation
of tyrosines and sugars; a function of the Golgi
modifying enzymes
markers for portions of Golgi stack
protein sorting in Golgi
performed for the next step so proteins are ready: plasma membrane/extracellular space, endosome/lysosome, peroxisomes, etc.
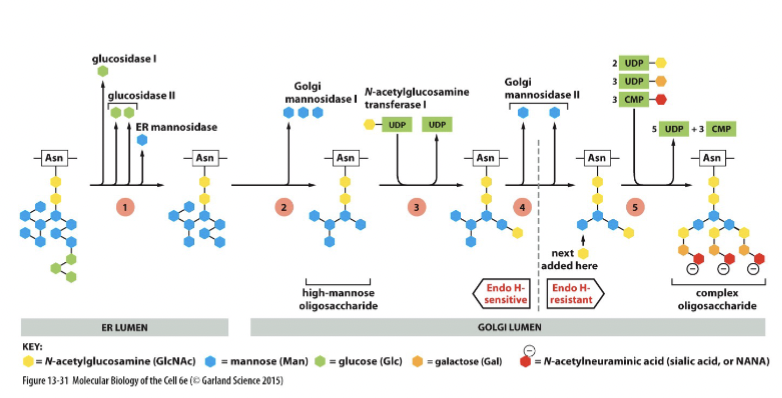
Endo H
bacterial protein (experimental system) that removes entire tree of N-linked oligosaccharides. It is blocked by oligosaccharide modifications in late Golgi
After processing in medial Golgi, glycoproteins become resistant to hydrolysis by this enzyme. Can use this resistance to track progression of the glycoprotein through the Golgi stack.
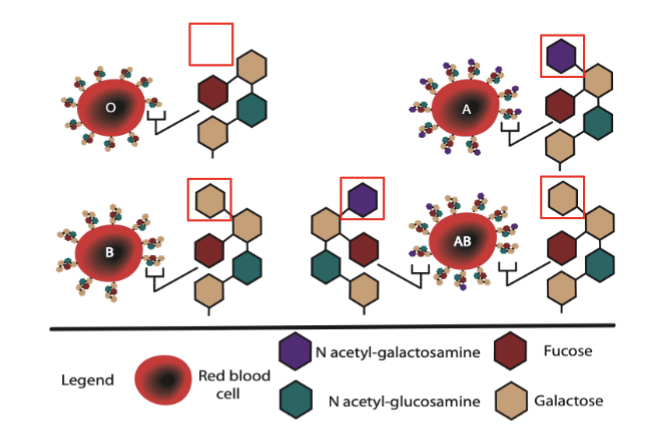
ABO blood type
example of glycosylation.
Genetic basis for controlling this: sugar transferase has two alleles, N-acteylgalactosamine transferase (A) and galactosyl transferase (B) that differ by only 4 aas, and the allele inherited determines the blood type - has different types of sugars present on membrane to distinguish type.
transport Golgi to plasma membrane
includes constitutive and regulated exocytosis. The vesicular contents released into extracellular space, and the vesicle membrane fuses to the plasma membrane
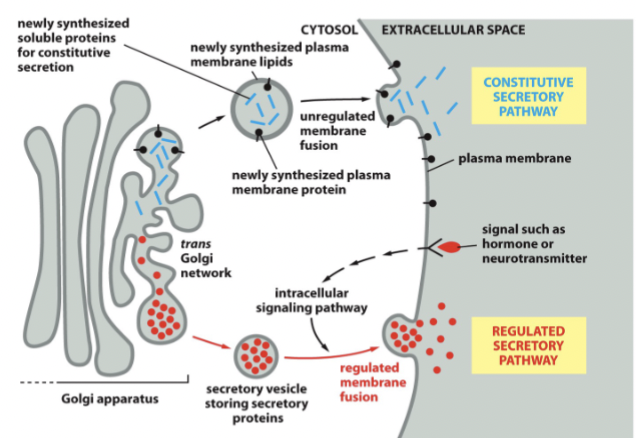
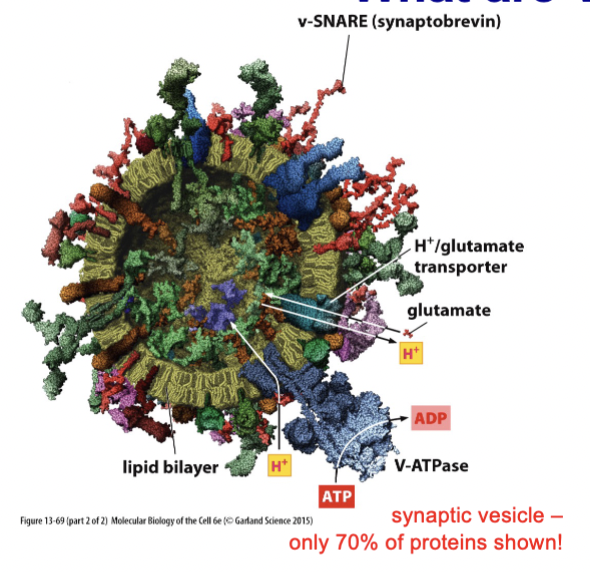
synaptic vesicles
small structures found in neurons that store neurotransmitters such as acetylcholine, serotonin, and glutamate, and are involved in the exocytosis process at synaptic connections, facilitating communication between nerve cells
Membrane fusion is often regulated as SNAREs interact and tie these (pre-docked) to membrane, yet don’t fuse because fusion only occurs after recognizing action potential
chemical characteristics of glycosylation
rigid, protrudes away from the protein or membrane to act as a sort of protective shell
aid protein folding: rigid structure limits peptide movement
highly hydrated (attract water molecules to act as a cushion in the EC matrix)
can provide protective “coat”
can add negative charge if sulfated
biological functions of glycosylation
resistance of extracellular matrix to compressive forces
membrane protection from hydrolysis
viral membrane binding (hemagglutinin)
blood type