BIO 205: EXAM 1 CONTENT
1/93
Earn XP
Description and Tags
FREEDMAN
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
94 Terms
Nuclear Pore Complex
A protein structure that serves as a channel between the cytosol and inside of the nucleus. Each pore is comprised of 30 proteins (~)
They serve as gates; responsible for selective permeabillity
Nucleus
Center of cellular data/info
enzymes interact with key centers to produce RNA
mRNA is produced here, and is sent outside of the nucleus in order to produce proteins
Nucleolus
Site of ribosome assembly.
Ribosomal RNA binds to proteins to form ribosomes
Nuclear Envelope; what comes inside?
Nucleoside Triphosphates (NTPs; building blocks for DNA and RNA)
Proteins responsible for making DNA, RNA and ribosomes
NLS: What is it? Purpose? Composition?
NLS = Nuclear Localization Signal. It is a 17 amino acid sequence. During synthesis, if a protein is bound for/has a function in the nucleus (making it a nuclear protein) it will be synthesized with an NLS.
The Endomembrane System
The ER, Golgi, and membrane structures synthesized by them
Processing of proteins destined for secretion (Steps)
Protein Synthesis begins when mRNA delivers instructions to a Free ribosome. The ribosome synthesizes an ER signal sequence (go to ER)
SRPs (Signal Recognition Particle), which are abundant in the cytosol, bind to the ER signal sequence. This terminates synthesis.
The signal complex moves to the RER, where the SRP binds to its corresponding SRP receptor.
Binding to the receptor releases the SRP.
The protein is sent to the ER lumen where it continues to synthesize to completion. The ER signal sequence is removed.
How do proteins get to the lysosome for recycling?
Receptor Mediated Endocytosis
Macromolecules bind to the outside of the membrane surrounding the lysosome. a conformational change results in a membrane piece folding in and pinching off to create a vesicle while simotaneously encapsulating the molecule. The molecule is dumped into a early endsome. It has H+ Pumps that acidify the molecule. This causes the receptors to unbind and become recycled. As it acidifies; it becomes a late endosome. This then turns into a lysosome.
Phagocytosis
The membrane of a cell engulfs a molecule; forming a phagosome. The phagosome is delivered to the lysosome. They fuse and the phagosome is digested.
Autophagy
Same concept as pahgocytosis except its damaged organelles or other dysfunctional parts of the cytoplasm are marked, has a vesicle formed around it, and digested by the lysosome.
Cytoskeleton
Dense, complex network of fibers. Maintains cellular shape via structural support.
The cytoskeleton also lets the cell:
Alter its shape
Shift its contents
And locomote
3 components of the cytoskeleton
Actin filaments (micro)
Intermediate filaments
Microtubules
Actin Filaments
Smallest in terms of diameter; AKA microfilaments
They are made of actin; a globular protein subunit. (globular = folded compactly; like a ball). Very abundant; 5-10% of the total protein in an animal cell.
Functions:
Myosin binds to and hydrolizes ATP which changes the shape of the actin head and results in a pulling on actin creating movement. This movement is involved in :
Cytokinesis; splitting of cells
Cytoplasmic streaming: basically the controlled circulation of cytoplasm (and thus organelles) in the cell. Helps transport materials
Cell crawling: extension and retraction of actin results in movement
Maintains cell shape by resisting tension
participates in muscle contractions
Intermediate Filaments
There are different kinds of these fibers; 70 genes code for variants. Most are keratins.
These filaments have identical ends and do not aid movement; they exist for structure of the cell and they also anchor the nucleus and other organelles.
A type of intermediate filaments you might wana know:
Nuclea laminas. They give the nucleus its shape, anchor chromosomes, staballizes the nuclear envelope. They break up and reform during and after cell division.
Microtubules
Made of alpha and beta tubulin that form dimers via noncovalent bonds. These dimers keep forming to make a protofilament; which is a long hollow tube. Since they always align in a certain way (alpha binds with beta) there will always be an alpha at one end (minus end) and a beta at the other (plus end). These dimers polymerize in this orientation.
MTOC synthesize these. The plus ends grow outward of the MTOC (usually somewhere near the nucleus) The MTOC is synonymous with centrioles.
Their functions are alike actin filaments. They maintain shape by resisting compression and provide a sort of scaffolding blueprint for organelles.
They move cells via flagella or cillia
They move chromosomes during cell division
They assist the formation of the cell plate during cell division
They provide “tracks” for intraceulluar transport
Actin:Treadmilling
Actin has a + and - end because it is composed of subunits. Subunits generally add more to the + end and add less to the - end. Specifically ATP-ACTIN binds more readily to + end. Over time, the ATP on ATP-ACTIN hydrolyzes, which is less stable and tends to dissasociate which is why the - end is the way it is.
This whole thing is called treadmilling. This is responsible for some movements.
Microtubule Stabillity
The - end of a microtubule is typically anchored in a MTOC. The action occurs at the + end
At the + end, subunits composed of alpha beta tubulins in the form of a dimer. They come with GTP attatched to the beta tubulin. The beta tubulin hydrolizes the GTP. This creates a high energy structure that collapses quickly. However, the microtubule generally stays together as long as there is a GTP cap on the + end. If hydrolysis of GTP occurs on the last dimer of the + end, catastrophy occurs, and the tube falls apart.
The defining difference of Eukaryotes vs prokaryotes??
Eukaryotes have membrane-bound
nucleus
Prokaryotes lack membrane-bound
nucleus
What kind of cells do organisms of the Archaea phylogeny possess??
Prokaryotic
What are some things prokaryotic and eukaryotic cells have in common
At least one DNA chromosome
ribosomes
a plasma membrane made of a phospholipid bilayer
cytoplasm (so organelles and cytosol)
cytosol (jelly/liquid stuff)
Prokaryote: Location of a large, circular DNA molecule? What things help organize its structure and function
Nucleoid. Associated with regulatory proteins
Most prokaryotes have this protective extracellular layer. What is it. What does this thing do.
Also; do eukaryotes have this… thing?
Cell wall. A tough rigid layer that surrounds the plasma membrane and holds the shape of the cell and maintains rigidity. Since water enters prokaryotes via an ionic gradient; the volume becomes high and the associated pressure is resisted by the cell wall.
Some eukaryotes do! plants have them. Not animals though
Flagella
An assemblage of proteins that is powered by a molecular motor. It allows for movement of the cell.
Fimbriae
Needlike projections that extend from cells and allow attatchment to extracellular objects
Can Eukaryotes be unicellular
yes
Function and advantage of organelles?
Organelles compartmentalize volume within the cell. This:
Seperates incompatible chemical rxns
Increases the efficency of said reactions (Substrates can be made readily available in high concentrations, enzymes can be highly concentrated
Nucleus: some characteristics abt membrane?
Double membrane nuclear envelope
pore like openings
Inside surface bound by nuclear lamina
The part that is NOT the nucleolus iscalled chromatin
Nucleolus
Region inside the nucleus that produces rRNA that is used to produce ribosomes. Is also the site where ribosomes are produced in general.
Ribosomes
Units that produce proteins. Are either present on the surface of the RER or free in the cytosol. Proteins that are produced on the RER are typically bound for secretion. Proteins made in the cytosol are typically bound for organelles or exist free in the cyotosol.
Define the area of the nucleus that is NOT the nucleolus
Chromatin
Endoplasmic Reticulum
The ER is an extension of the nuclear envelope. It has two regions;
The RER which is studded with ribosomes on the outside that produce proteins that enter the lumen of the ER where they undergo more folding and processing (RER produced proteins are extracellulaaly bound and end up as things like messages to other cells or membrane transport proteins). After processing they are packaged into vesicles and transported to their destination
The SER catalyze lipid reactions with its enzymes to either synthesize or degrade lipids or threatening molecules. the SER also stores Ca2+.
NOTE: some proteins actually are shipped to some organelles from the RER
Golgi apparatus
Contains membranous compartments called cisternae. The cisternae on the Cis side (facing nucleus) recieves (via merging) vesicles containing RER proteins and the trans side ships them to organelles or out of the cell. Each cisternae’s function is related to the enzymes it contains. As it moves thru the golgi, it will be processed (labels added for destination) by enzymes and packaged for delivery
Cargo moves from cis to trans.
Lysosomes
Contains enzymes that hydrolyze different kinds of molecules for purposes of recycling. It does this with acidic chemical reactions (ex proton pumps).
Define the endomembrane system and its purpose
Lysosomes, Golgi apparatus, and ER make up the endomembrane system. The system produces, processes and transports proteins, carbs, and lipids.
Vacuole
Vacuoles have various roles. They store water, contain hydrolases to digest and recycle macromolecules. Notice the simularity to lysosomes. They also store K+, Cl- and other solutes that draw in water.
Peroxisomes
Center for Redox reactions. Comes from empty ER vesicles. Often produces H2o2 which is then enzymatically broken down.
Mitochondria
Produces ATP. Has two membranes. Outer membrane is the surface of the organelle. The inner membrane forms sacs called cristae. There is a solution inside the inner membrane is called the mitochondrial matrix. enzymes and machineary used to make ATP are free in the matrix or embeded in the inner membrane. They have their own DNA (mtDNA) that are independent of the nuclear DNA. These code for things like mitochondrial ribosomes that make mitochondrial proteins. Yet most proteins for mitochondrial function come from the nucleus.
Chlropoplast
photosynthesis. These chloroplasts have 3 membranes; innermost = sacs called thylakoids stacked to form grana, which are then surrounded by stroma. Chloroplasts also have their own DNA.
Endosymbiosis theory.
chloroplasts and mitochondria were once bacteria that were engulfed by eukaryotes.
Cell wall in fungi, algae and plants.
They have it. its similar to bacterial cell walls.
How do animal cells have structural support and protection if not by cell walls?
a secretion of proteins and polysaccharides called the extracellular matrix. Rods or fibers run through the matrix. Can also be used to attatch themselves to each other.
Nucleus of an atom
protons (+1) and neutrons (0). electrons are orbitals (-). Protons and neutrons in equal numbers = neutral.
Elements
single kind of atom. define by # of protons (because nuclear charge determines the element).
Atomic number
number of protons in a atom
mass number
sum of protons and neutrons (which have a mass each of 1 dalton)
Isotope defined by?
number of neutrons (extra neutron makes it heavy)
Heres a good example of how neutrons and protons work to define an atom and its mass.
Example: All carbon atoms have 6 protons:
– Carbon-12 has 6 neutrons; atomic mass 12 D a
– Carbon-13 has 7 neutrons; atomic mass 13 D a
– Carbon 14 has 8 neutrons; atomic mass 14 D a
– FYI: Carbon 14 used to date artifacts. Undergoes
radioactive decay by ½ every 5 thousand years vs
carbon 12 which basically does not decay
what are the 99% of atoms in da bod?
C, H, N, O, P and S
e- shells
Electron shells:
– Each electron shell contains specific number of
orbitals
– Electron shell comprising single orbital can hold
up to two electrons
– Shell with four orbitals can contain up to eight
electrons
Shells fill from inside to outside
Valence shell
outermost e- shell. intermolecular interactions occur within these electrons.
Atom’s valence
number of unpaired electrons. determines # of possible bonds.
When is max stabillity of an atom achieved?
a full valence; achieved by chemical bonds.
Covalent bond
sharing of electrons; makes a molecule.
electronegativity
strength of which electrons are pulled to an atom. results in the polarity of a bond.
is water polar. what happens if it is
yes. it h bonds with itself and other polar molecules.
properties of water as a result of h bonding/ polarity
Excellent solvent (anything charged or polar will interact with waters polar parts and will thus stay in solution). H- Bonding makes it possible for any polar molecule to dissolve in water.
2. Cohesive and adhesive properties
3. Solid (ice) is less dense than liquid (ice floats)
4. High thermal capacity (specific heat and heat of
vaporization – takes a lot of energy to change temperature)
Why do non polar molecules attract each other (and thus cluster away from water and other polar molecules)
van der waals interactions
Cohesion
attraction between like molecules. Water is cohesive cause it sticks to itself.
adhesion
attraction between different molecules. water is adhesive because it sticks to polar or charged stuff.
Water and density and the weird thing about it.
Most substances shrink as they turn to the solid phase, yet water expands as it freezes. it is denser as a liquid than as a solid. This is because of the strength of its H bonds.
Waters heat capacity.
many h bonds must be broken. so its high. again, because of strong intermolecular (hbonding) forces.
Since water has such strong intermoleculer forces it vaporizes at a very high heat.
acid
give up h+. less than 7 ph
base
takes H+. greater than 7 ph.
pH
power of hydrogen
buffer
minimize ph changes. Buffers help maintain homeostasis, relatively
constant conditions, in organisms
– Add H+, buffer pulls some out of solution (counters the
addition of H+)
– Subtract H+, buffer adds more to solution (counters
the subtraction of H+
carbon.
almost all molceules in the bod have carbon. makes 4 bonds
Organic compounds—molecules that
contain carbon bonded to other elements:
– Limitless array of molecular shapes
– With different combinations of single and
double bonds
Amino groups
attract protons cause its basic
carboxyl groups
are acidic
carbonyls
sites for linkage of more complex molecules
hydroxyl
polar and hydrophillic
phosphate groups
2 negative charges
Sulfhydryl groups
makes disulfide bonds
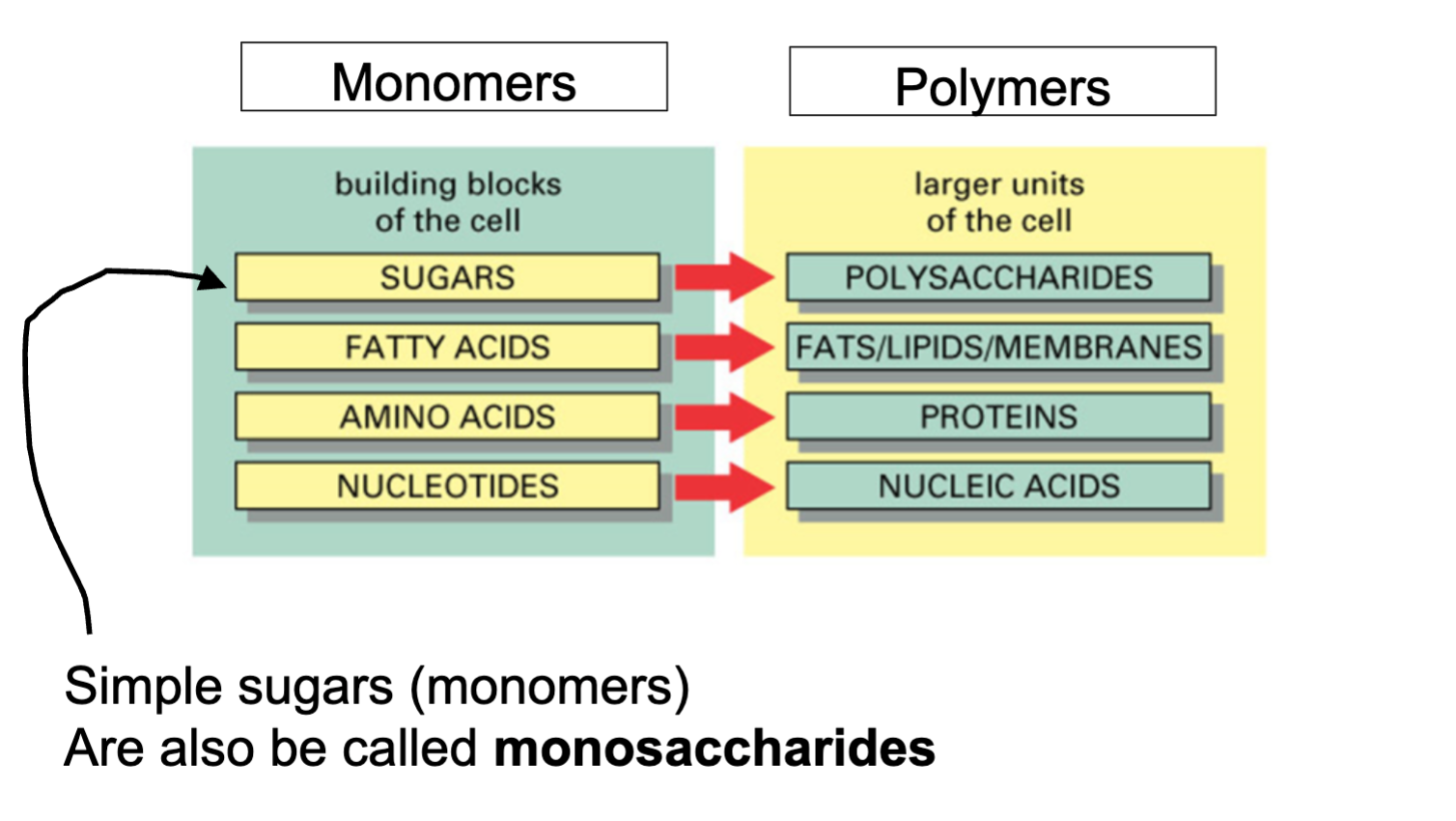
Monomers
Molecules used as subunits to create bigger molecules
ex monosachrides
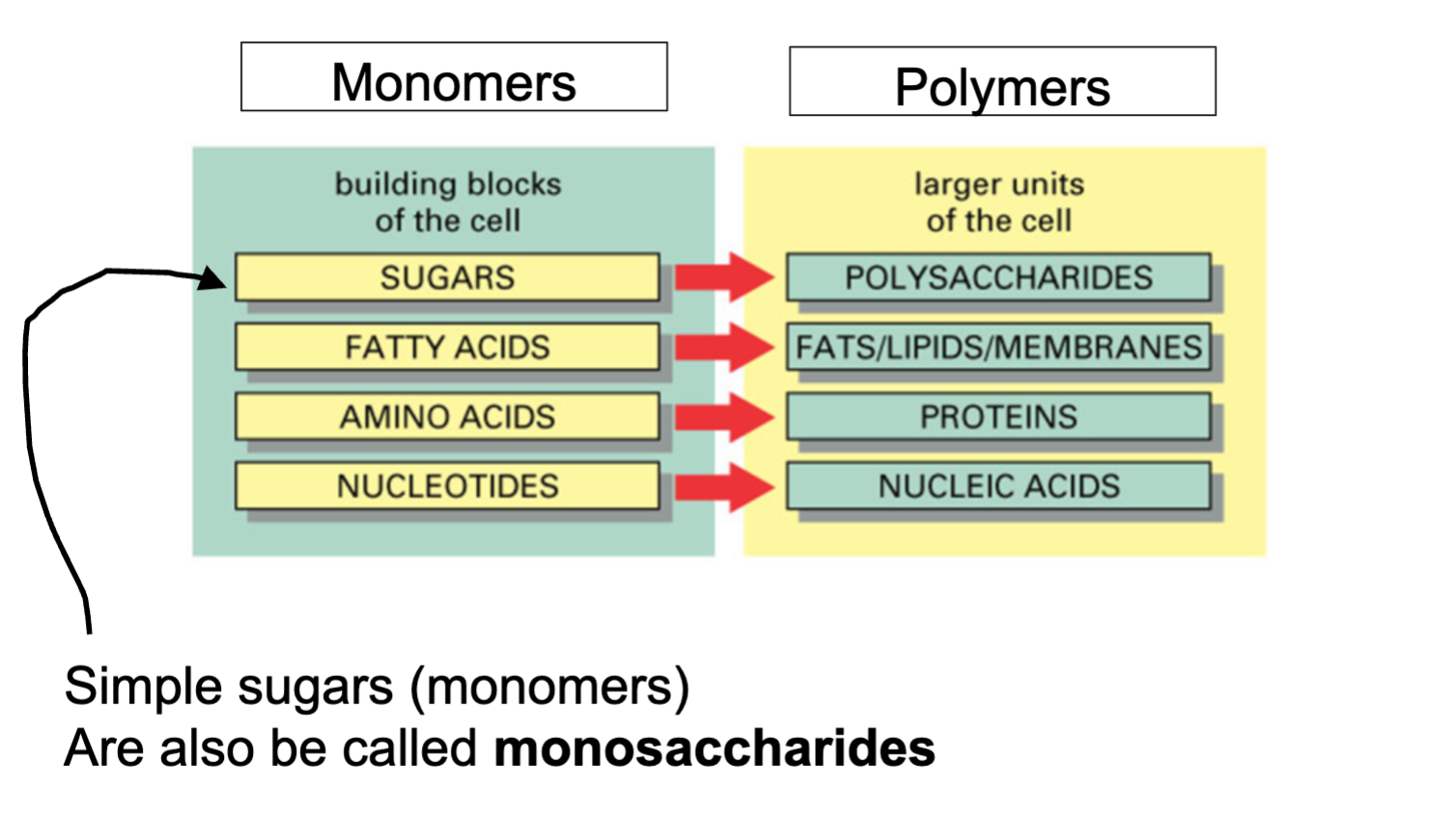
polymers
large chain molecules that consist of monomers
ex; polysaccharides
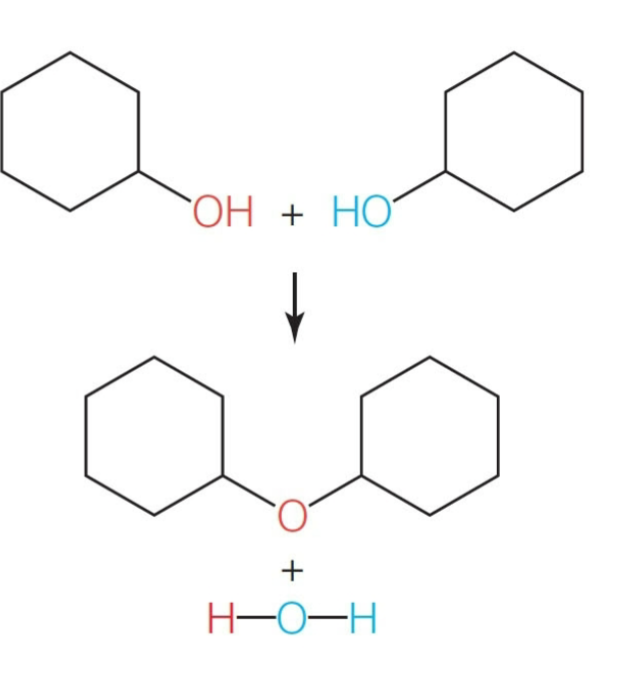
What reaction creates a bond between two molecules?
Condensation reaction. water is produced as a byproduct. As you can see all atoms are present after the reaction; an ether is formed to dimerize the molecules and h2o is shown as another product as it functioned as the leaving group.
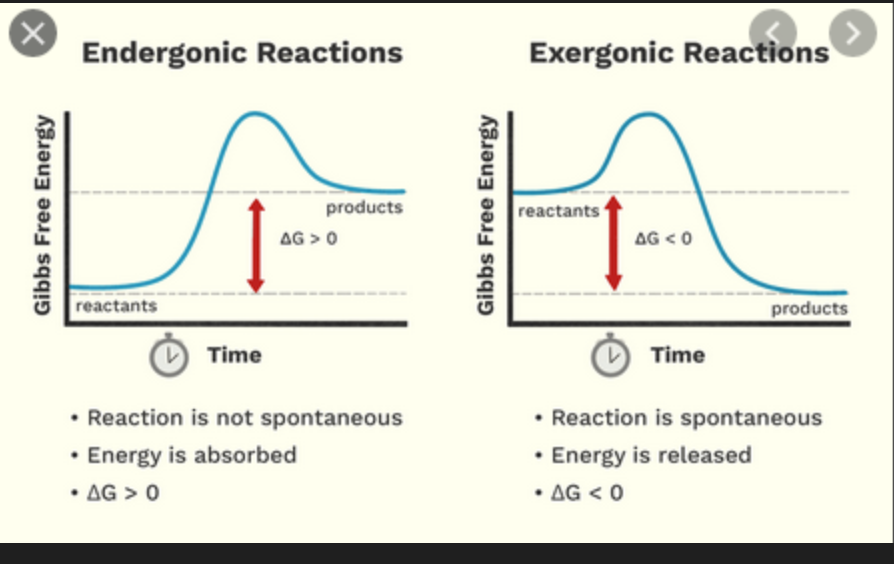
A condensation reaction requires an input of energy to form monomers. Is this reaction exergonic or endergonic
endergonic.
If energy must be put into the system; it doesn’t happen spontaneously, theorefore we must “push the reaction uphill” to a higher energy state, creating a + ^G
What reaction breaks a bond between two monomers
Hydrolysis. Essentially the inverse of condensation. If you add water in an influx, it will be consumed to break the bonds
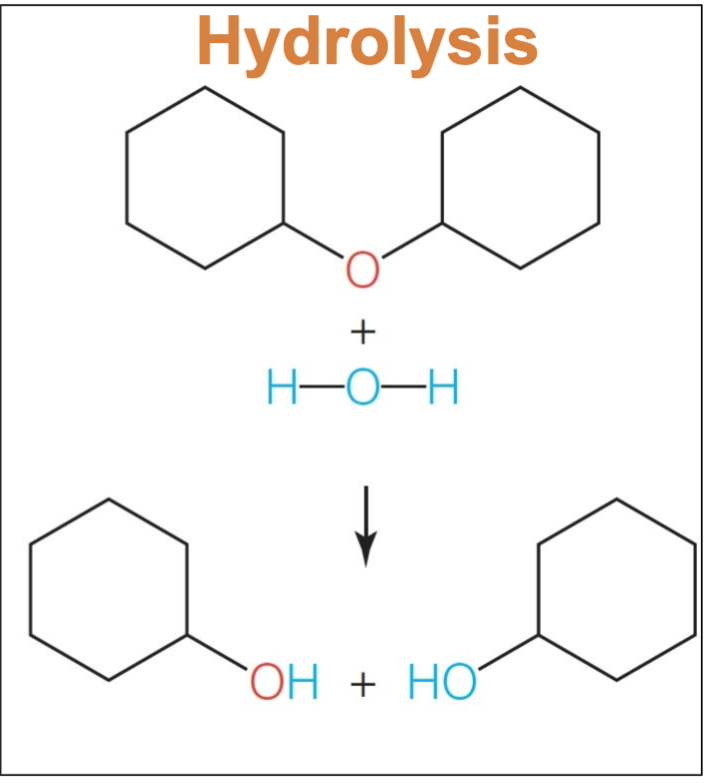
A hydrolysis reaction happens sponatneously. What is the delta G of the reaction?
negative.
In what direction are amino groups added to a string?
From N terminus to C terminus.
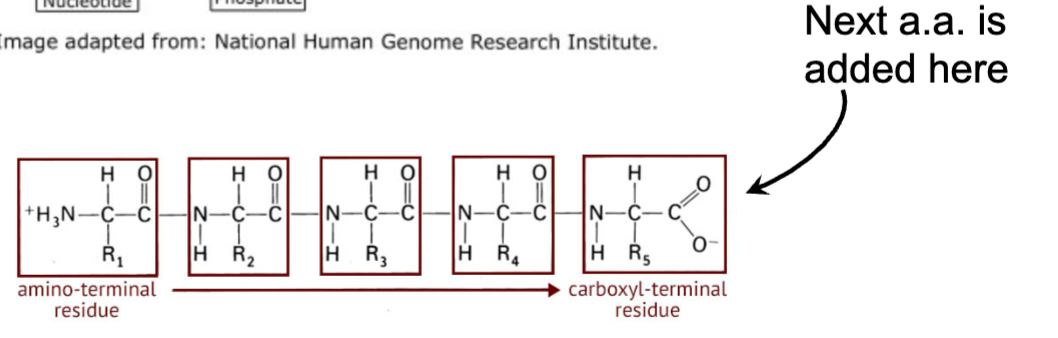
in what direaction are amino acids added to a string
from 5’ (phosphate group bonded to the 5 carbon on the ribose) to the 3’ (additional group is added via a condensation rxn between the OH on the 3’ and the OH on the 5’)
Carbohydrate/sugars
fromula (CH2O)n
highly soluble
metabolized for energy
part of cell structure
used to identify cells\
Can be;
monosaccharides
disaccharides
oligosaccharides
polysaccharides
Glucose
Monosaccharide
Sucrose
Diassachride dimers of glucose and fructose
Lactose
diassachride dimer of galactose and glucose
Starch
Storage of carbohydrates in plants. Glucose monomers
Glycogen
storage of carbs in mamallians. Glucose monomers
Cellulose
Carbohydrate structural support in plants. Humans cannot break it down because of H bonding between neighboring chains
Chitin
Carbohydrate structural support in fungi and exoskeletons in insects. Strengthened via H bonding.