BIOL 200 Lecture 10: Chromosomes
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
32 Terms
In what stage of the cell cycle is DNA highly compacted, and how?
During metaphase, DNA is highly compacted into chromosomes so it can be transmitted to daughter cells.
In most stages of the cell cycle, what state does DNA exist in?
Chromatin
Extended form of chromatin
“Beads on a string” model
Nucleosomes are the beads, with DNA being tightly wound around proteins known as histones.
147 base pairs of DNA wrap around a histone, making 2 full turns
Histones
Positively charged proteins that negatively charged DNA wraps around.
Linker DNA
10-90 base pairs that separate the nucleosomes
what makes up the protein core of a nucleosome?
2 copies of 4 types of histones (H2A, H2B, H3 and H4)
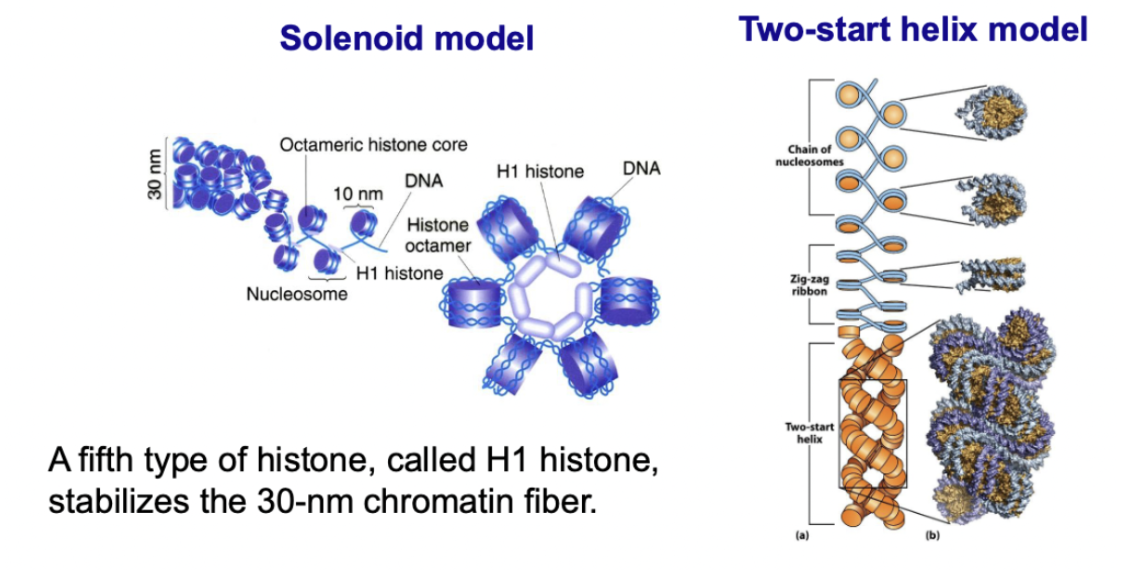
Condensed form of chromatin
fibre-like form of DNA
30nm in diameter
Multiple nucleosomes condense to form a fiber
2 models
Solenoid model
Two-start helix model
H1 histone stabilizes 30nm chromatin fibers by organizing them into particular structures
Histone tails
unstructured regions of histones (lack secondary structure)
stick out from histones
modified post-translationally, which regulates chromatin condensation
Histone tail modification
Post-translational
Acetyl group added to amino group on lysine, which neutralizes positive charge
Other modifications can change the charge of side chains, including methylation, phosphorylation, and ubiquitination
These changes are important to signal to the cell how compacted the chromatin should be.
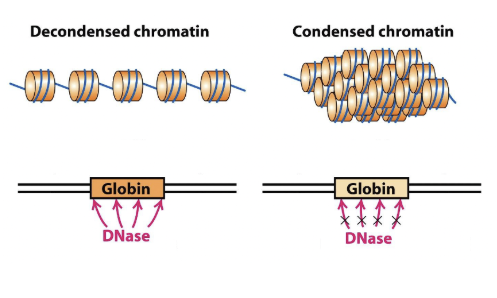
How does chromatin affect transcription?
Decondensed chromatin is accessible and can bind to other proteins
Condensed chromatin is protected by compaction and not available for binding.
This determines which genes are transcribed.
Polytene chromosomes
Several parallel, identical chromatids
Giant interphase chromosomes formed when multiple copies of DNA are repeatedly replicated without separation, allowing them to remain clustered.
They are found in certain tissues, such as the salivary glands of some insects, and are used to study gene activity and chromosomal structure.
Polytene chromosome puffs
show chromatin condensation with transcriptional activation
Active transcription sites on polytene chromosomes
Fluorescent microscopy can label active polymerase Pol II and differentiate it from its inactive form.
Heterochromatin
compacted regions of chromatin with nucleosomes (rich in repetitive DNA, poor in genes)
Euchromatin
decondensed regions of chromatin that are rich in genes and transcriptionally active, but poor in repetitive DNA.
Long chromatin loops
Non-histone proteins provide structural scaffold for long chromatin loops
rich in genes
1-4 million base pairs in length
Structural Maintenance of Chromosomes (SMC)
mediate DNA looping
Dimer proteins that contain a hinge domain, with ATPase head, and coiled coil domain.
50nm in size
bind to DNA, containing DNA inside ring shape and start moving to form loops. Loops interact among themselves, forming clumps that represent a chromosome.
Compaction during mitosis
Chromosomes occupy most of the nucleus during interphase
Each chromosome has its own territory
Compaction requires formatiob of loops, as DNA must be compact for separation.
Uses condensins
Condensins
specialized SMCs that condense chromosomes during metaphase
mitotic chromosomes are organized in loops around central core
Condensin 1 forms central scaffold with loops around it, while condensin 2 further compacts the loops into clusters of smaller nested loops
Compacts chromatin by 10,000 fold.
3 functional elements chromosome require for replication and stable inheritance
Origin of replication
Centromere
2 telomeres
Centromeres
Where chromosomes are linked to spindle microtubules during mitosis
microtubules are cytoskeletal structures that help pull DNA apart
Centromeric Protein A (CENP-A)
centromere specific histone variant contained in a nucleosome
Links to specialized protein complex called kinetochore, that connects centromeres to microtubules.
Telomeres
protective caps of repetitive DNA and proteins at the ends of chromosomes
protect the ends from exonuclease and end-to-end fusion
Contain simple repeat DNA sequences
derived from the loss of DNA during replication, ensuring chromosome stability.
TTAGGG in humans
TTGGGG in ciliate protist
What is the telomere problem?
Since the lagging strand cannot be completed because primers are needed and DNA cannot be synthesized, chromosones should shorten at the ends in each replication.
eventually this will lead to gene loss
Telomerase
A DNA polymerase that can extend telomeres, restoring chromosome length
Reverse transcriptase that carries its own template RNA complementary to the telomeric DNA repeat
What is reverse transcriptase?
DNA polymerase that uses RNA as a template (opposite of usual transcription)
How does telomerase work?
by extending the template strand as it is a reverse transcriptase, it gives primase more template DNA to prime on, preventing the shortening of the ends.
Where is telomerase active?
only in germ cells and stem cells (require replication several times)
somatic cells divide only a few times, so the telomeres they have without telomerase are enough.
It is also active in cancer cells, making it a target for cancer therapy.
Budding yeast experiments
done to understand chromosomal elements and help in figuring out what is needed to contain a piece of DNA in a cell.
How to find an origin of replication in budding yeast
Use a circular DNA molecule (ex. budding yeast) carrying a gene coding for an enzyme for the synthesis of leucine, which is transformed in mutant cells incapable of synthesizing leucine.
In cells that cannot produce leucine, no colonies are obtained
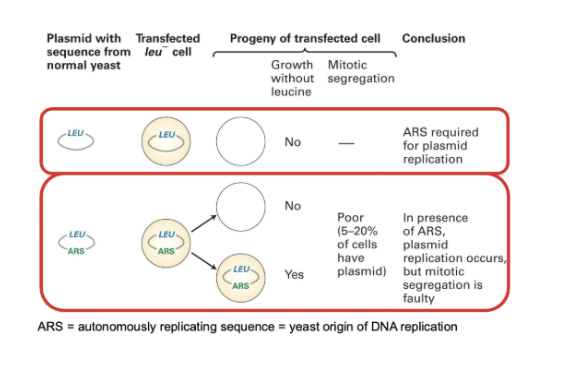
Autonomously replicating sequence (ARS)
yeast origin of DNA replication
If we have circular DNA with ARS and a marker and introduce them into cells that cannot produce leucine, we can begin seeing some colonies form. However, many will die as DNA is replicated, but not separated from other strands.
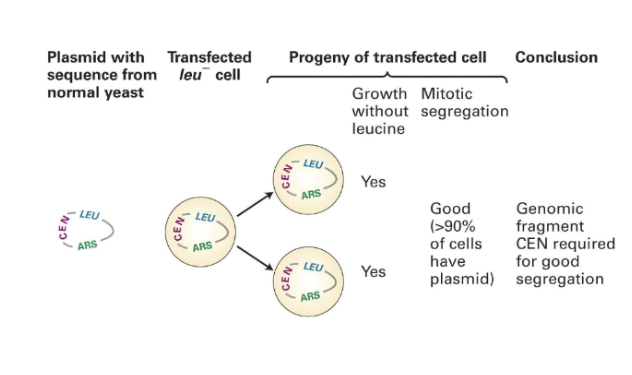
Introducing a sequence for the centromere in budding yeast (CEN)
CEN is a DNA sequence from a yeast chromosome centromere
we can introduce more pieces from yeast into DNA that already have the origin of replication, as well as a centromere.This allows circular DNA molecules to be efficiently transferred to daughter cells.
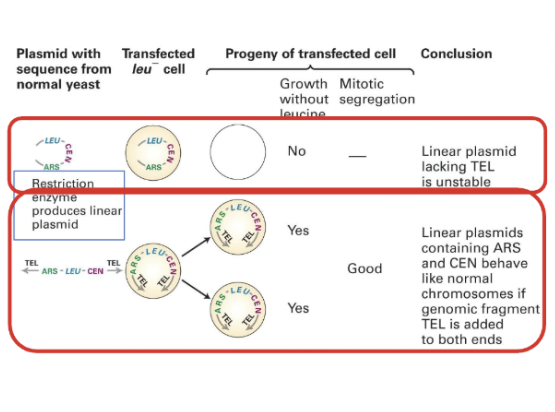
Purpose of introducing a sequence for telomeres in budding yeast
taking the circular DNA that contains the origin of replication and centromeres and cut it with restriction enzymes
introduces a linear plasmid. However, no colonies will form as linear plasmids without telomeres are unstable.
Only once a sequence for telomeres is added will the DNA be efficiently contained.
Linear plasmids
Linear plasmids containing ARS and CEN behave like normal chromosomes if genomic fragment TEL (telomeres) are added to both ends.