L5: Eukaryotic chromosome replication III
1/52
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
53 Terms
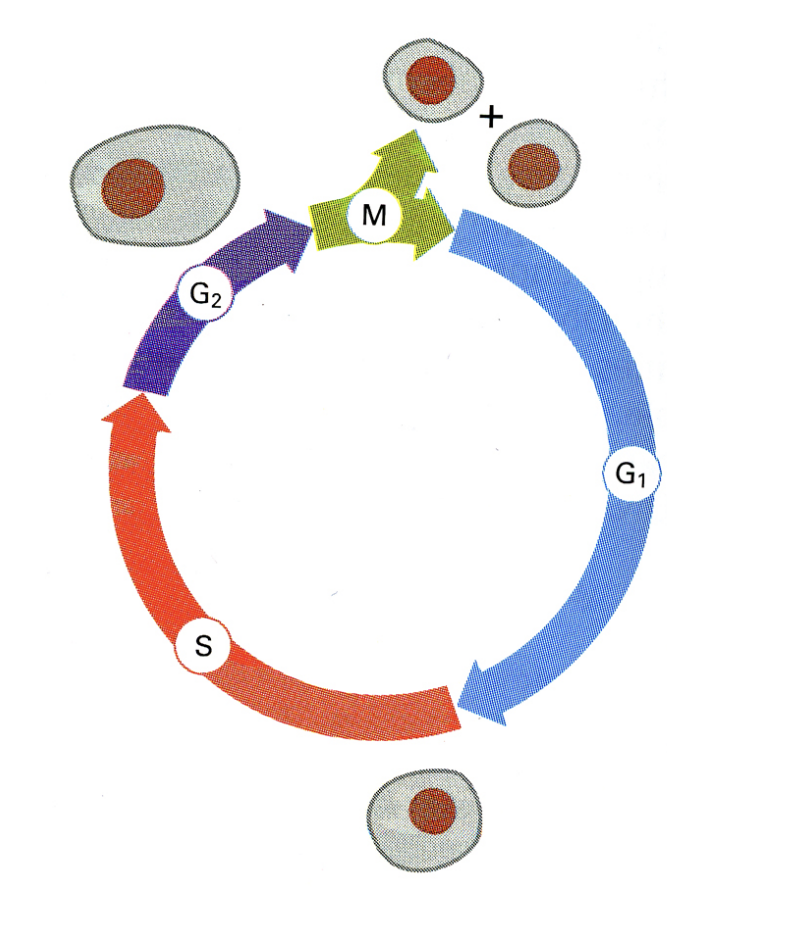
Cell cycle control of DNA replication:
DNA replication is tightly controlled during the cell division cycle
entire genome is replicated precisely once in S phase
separation of the replicated chromatids occurs in mitosis
BOTH EVENTS ARE STRICTLY SEPARATED
The licensing factor model: explaining how DNA is only replicated once
activator made
binds to DNA
bound activator stable and the free is destroyed
replication initiates at activator
activator destroyed by initiation or by fork
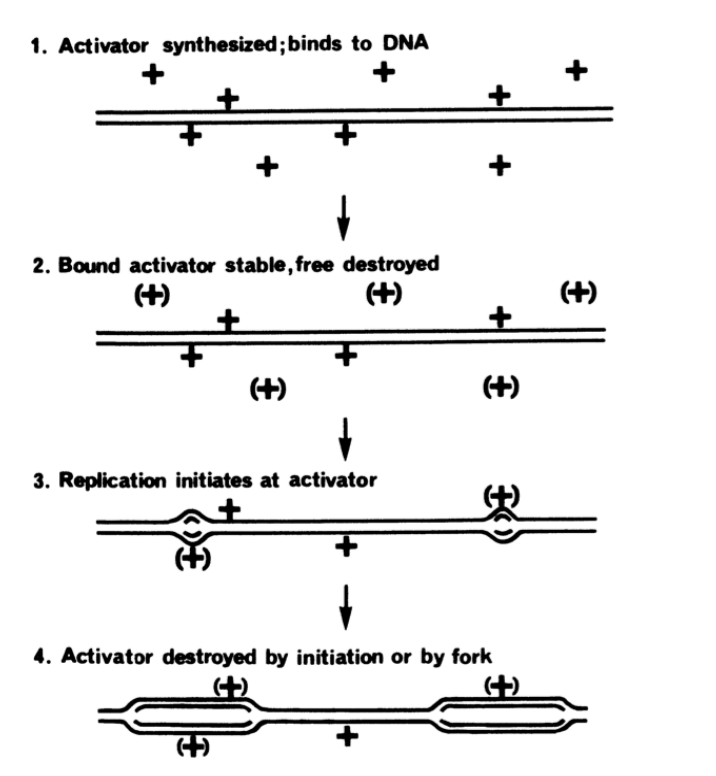
How re-initiation prevented?
helicase loading and activation under cell cycle control
What controls the start of DNA synthesis in eukaryotic cells?
cyclin-dependent protein kinase CDK complexes
→ kinases→ will phosphorylate the cyclins→ acts as an activation switch
These are related to
the complex of cyclin B and CDK1
that controls mitosis
Prime candidates iin vertebrates
cyclin A-CDK2
cyclin E-CDK2 complexes
What is also important:
Dbf4-Cdc7 protein kinase DDK
crucial for origin activation
and
initiation of DNA replication
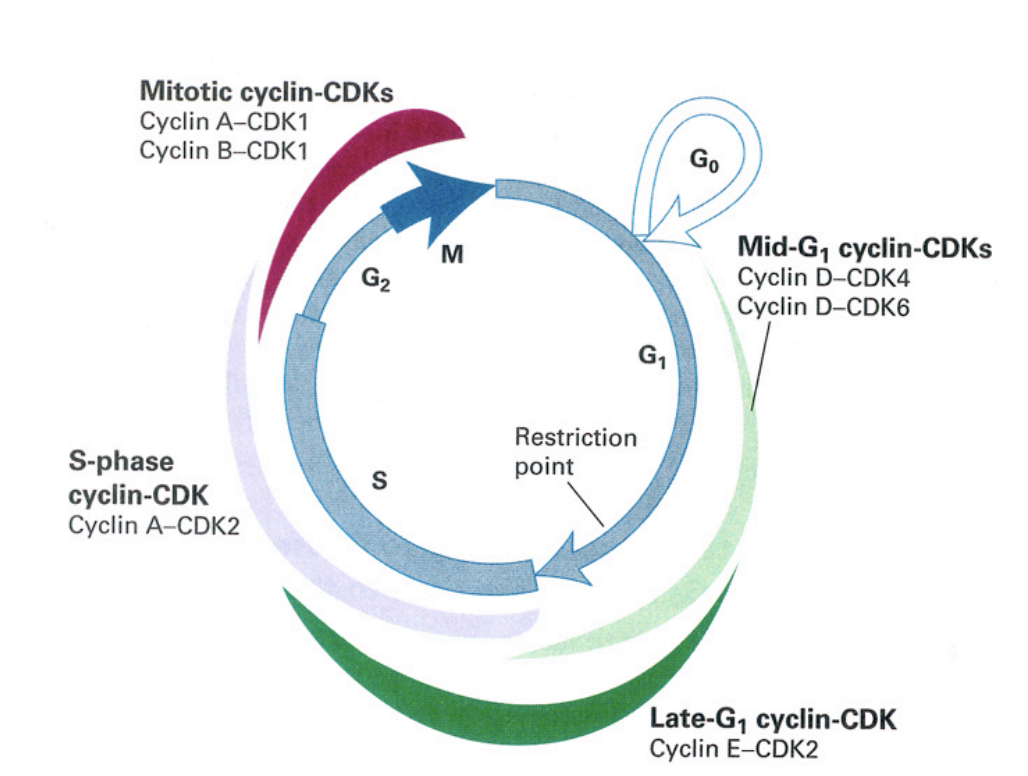
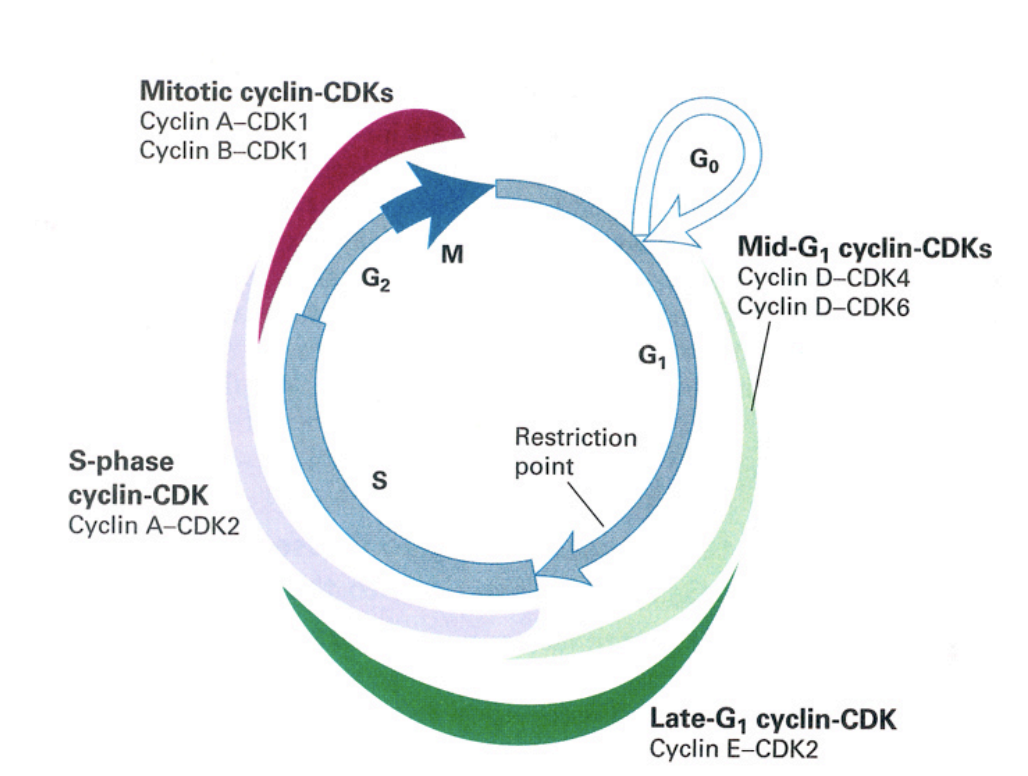
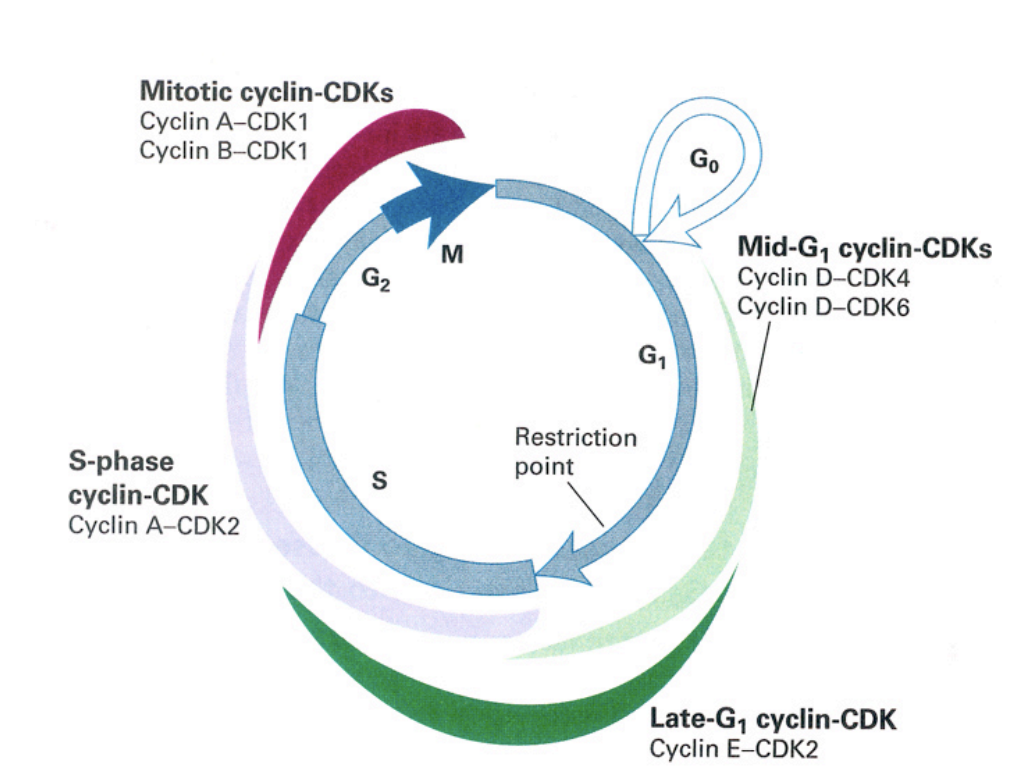
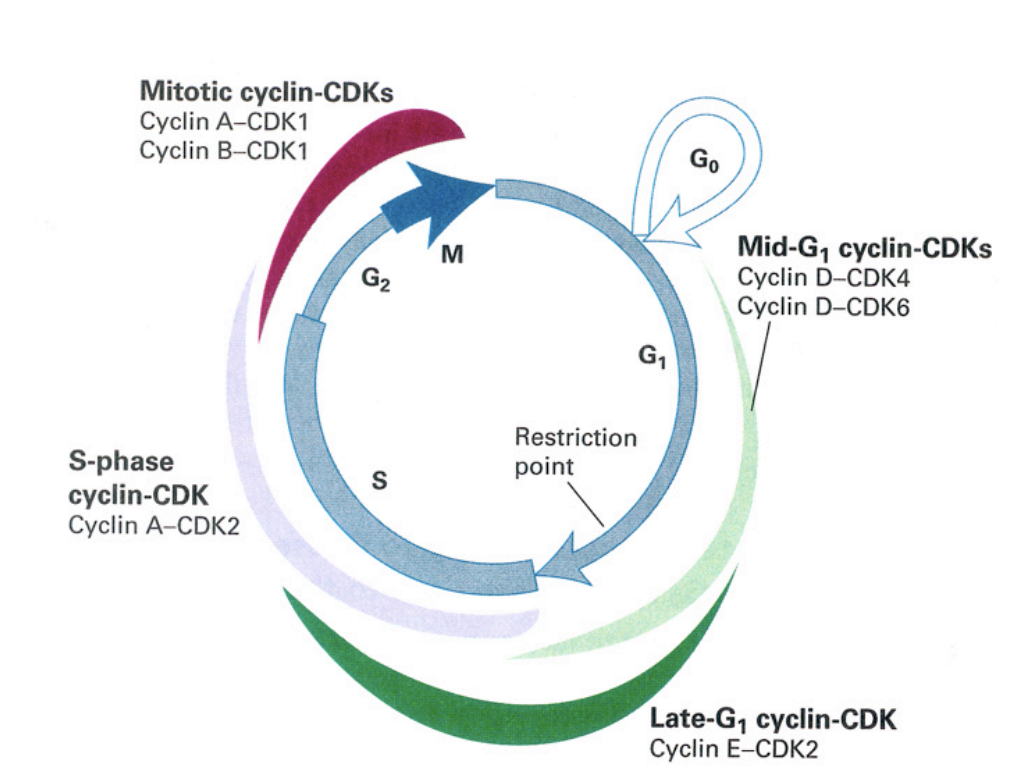
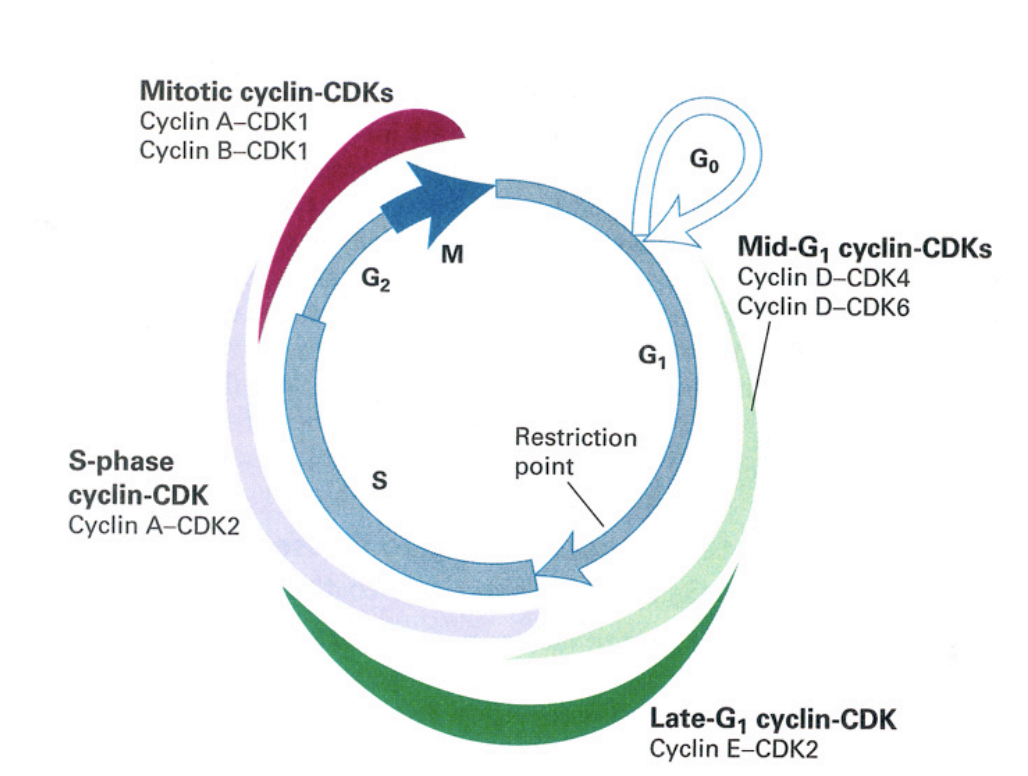
Cell cycle control machinery: cyclin expression
mid-G1 cyclin CDKs
cyclin D-CDK4
cyclin D-CDK6
late G1 cylin-CDK
cyclin E-CDK2
S-phase cyclin CDK
cyclin A-CDK2
mitottic cyclin-CDKs
cyclin A-CDK1
cycli B-CDK1→ very high especially
In order to ensure replication if only once per cell cycle, you need to be able to…
Initate replication only once per cell cycle
AND ALSO
Ensure that it is fully once
How to ensure that replication is FULLY once
many origins that are spread out
some are loaded
some are not loaded
stochastic nature
can be used as fall back if needed
Have many more than you actually need
Not only have many origins replicating at once→ so it is fully done and not too slow
Also→ have things to fall back on→ ensure that it will be all fully done
Once replication is initiated at an origin…
re-initiation is prevented
stops it happening twice in a replication fork
how…?
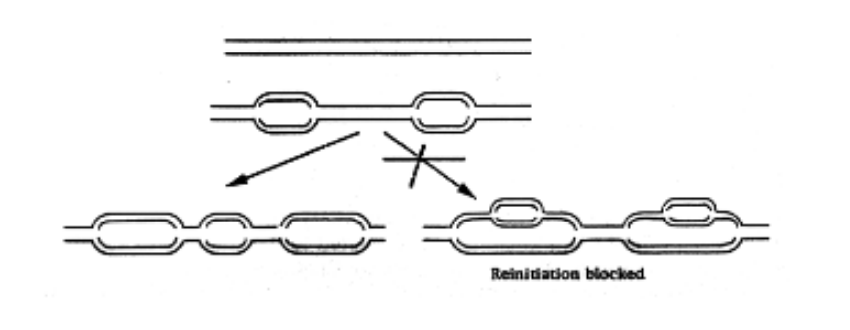
How is re-initiation of DNA replication prevented
at each origin is the essential pre-replication complex (pre-RC)
replication licence
→ assembled following exit from mitosis
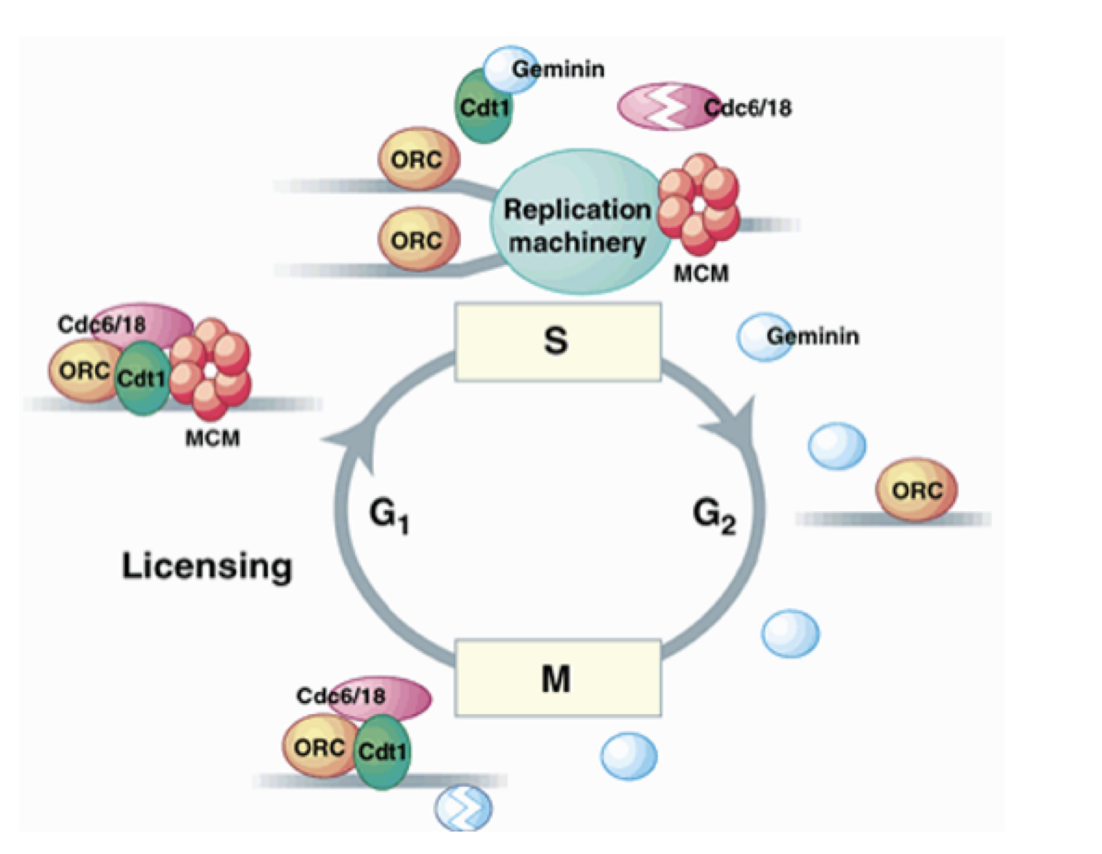
What is the licensing factor? (MODEL)
present at un-replicated chromatin at origins
required for origin activation/initiation
becomes inactivated during origin activation
absent on replicated chromatin until mitosis
i.e can only be licsensed BEFORE start→ coz environment changes other wise→ temporal and spatial separation between licensing and initiation
What fits this model:
MCM2-7/Cdt1 proteins fit the model best (seen above)
Oxidised double strand DNA also fits
MCM2-7/Cdt1 What does it consist of
ORC
cdc6
Cdt1
MCM (minichromosome maintenance) proteins
required for initiation
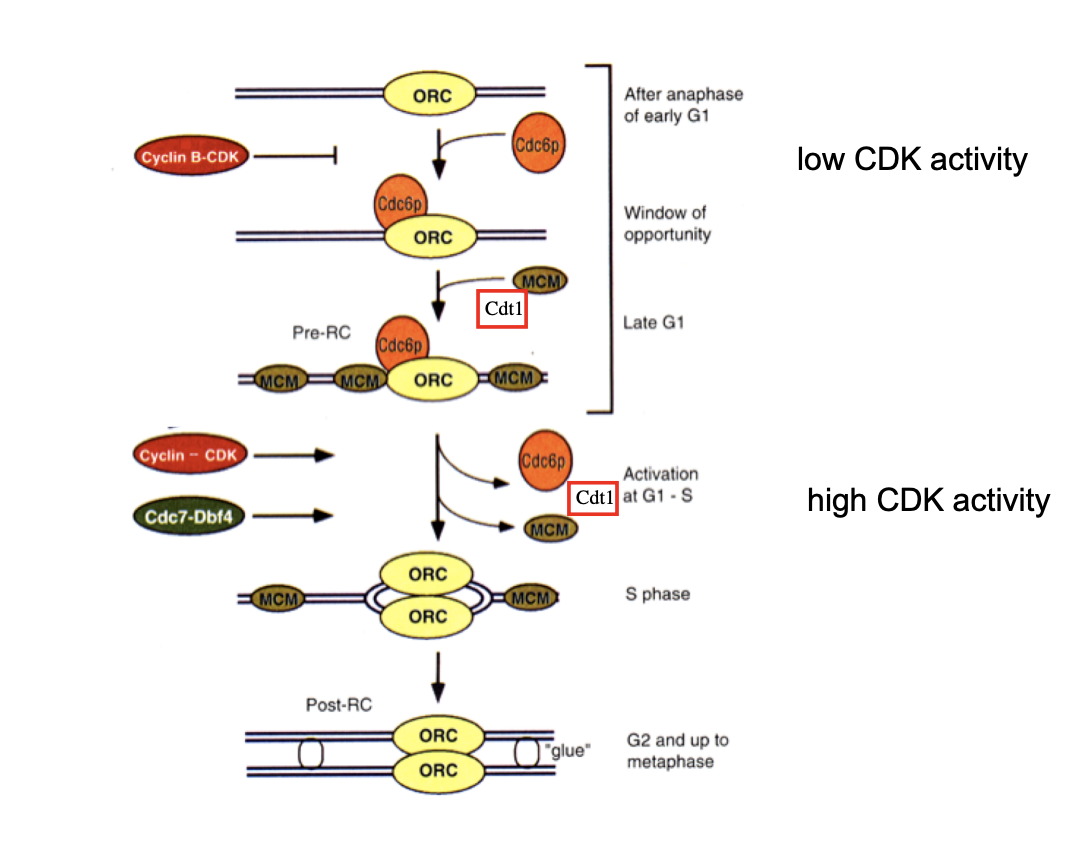
What happens after DNA replication is initiated
pre-RC is dismantled
Cdc6 and Cdt1 are degraded by proteolysis
MCM complexes are displaced from replication DNA
→ reformation of new pre-RCs and re-initiation of DNA rep are therefore prevented
until exit from mitosis
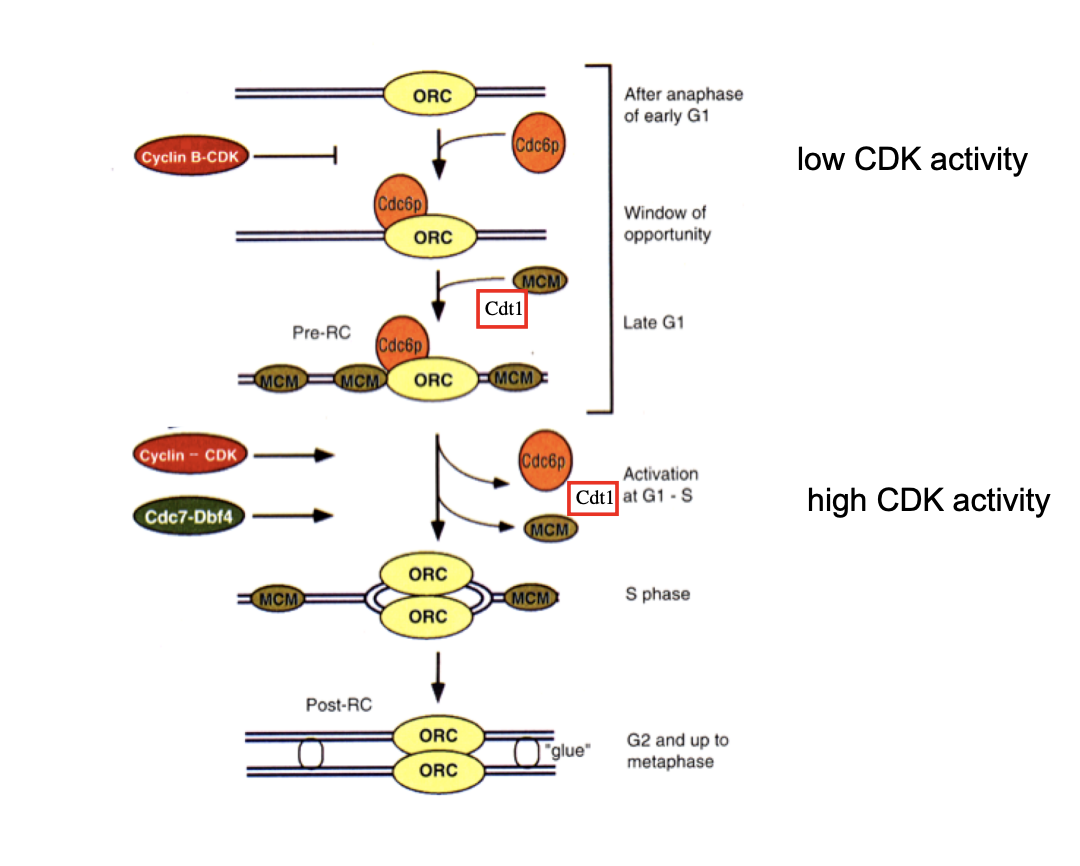
First level of control is exerted by CDKs:
High CDK activity is essential for origin firiing in S phase
and for preventing pre-RC re-assembly in S and G2
CDK activities are low in G1
This basic mechanism is conserved from yeast to humans…
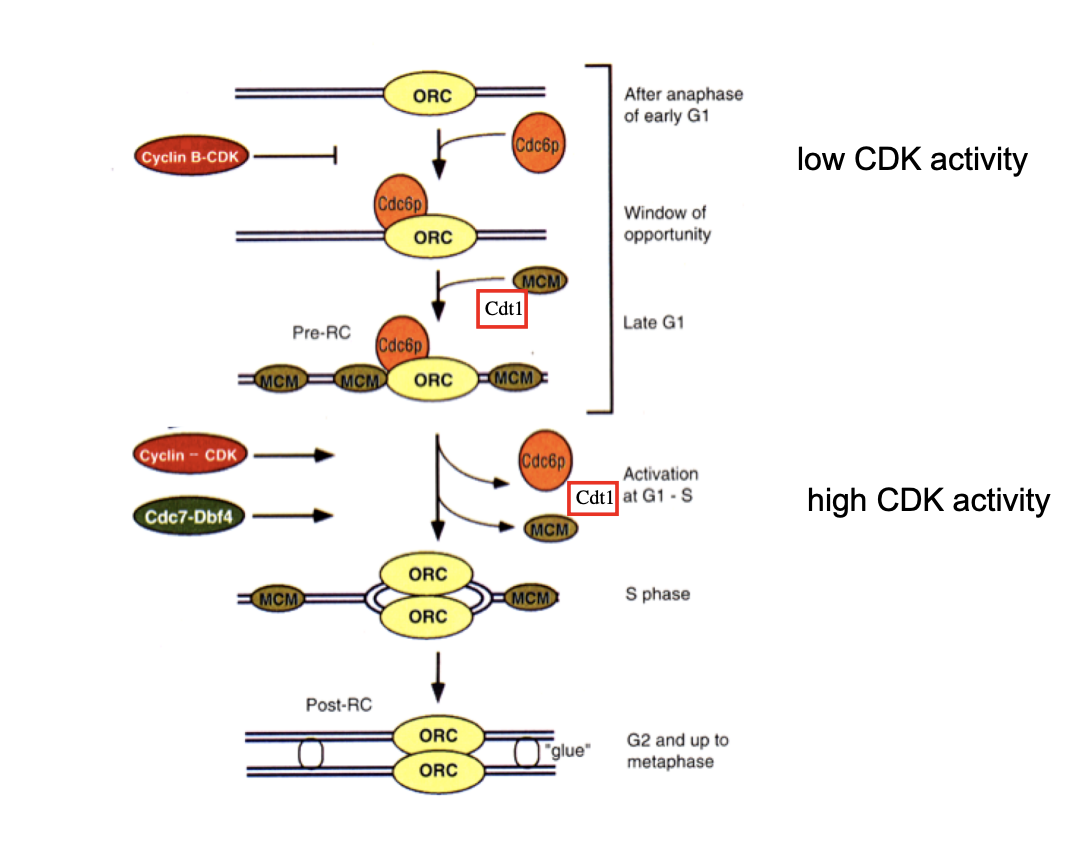
Second indpenendent level of control (found in mutlicellular organism) involves…
Cdt1
protein Geminin
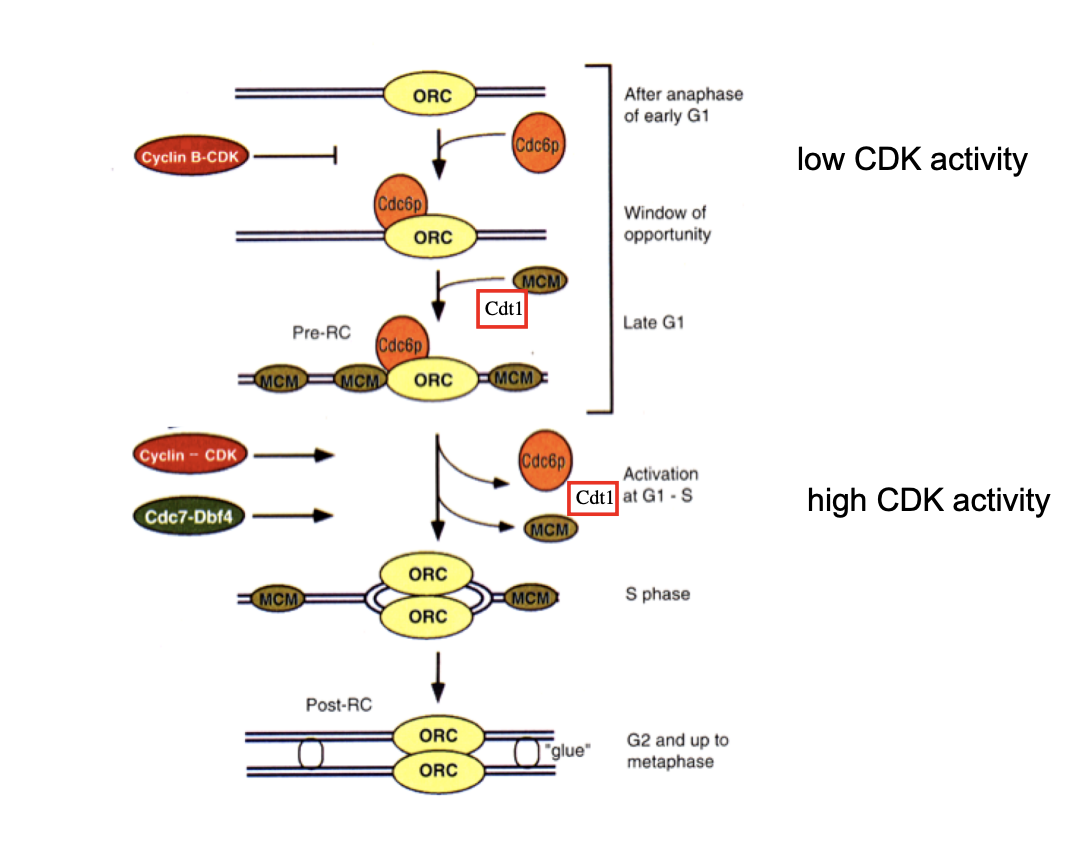
Geminin
binds to and inactivates remaining Cdt1 in S and G2 phase
prevents re-assembly of new pre-RCs after initiation of DNA replication
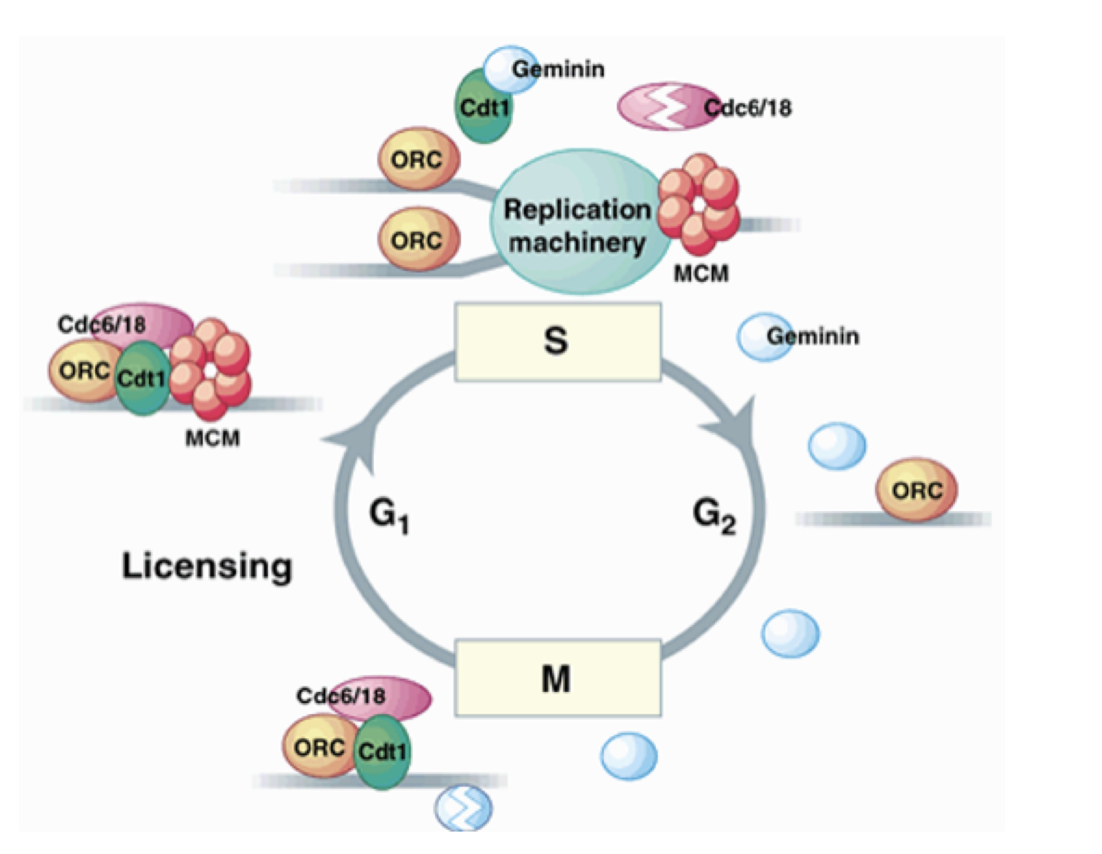
In mitosis, for the subsequent G1 phase…
Geminin is degraded (perhaps due to CDKs)
allows Cdt1 to assemble new pre-RCs
for the subsequent G1 phase
the degredation and remaking of Geminin must be more efficient than the making and degredation of cdt1
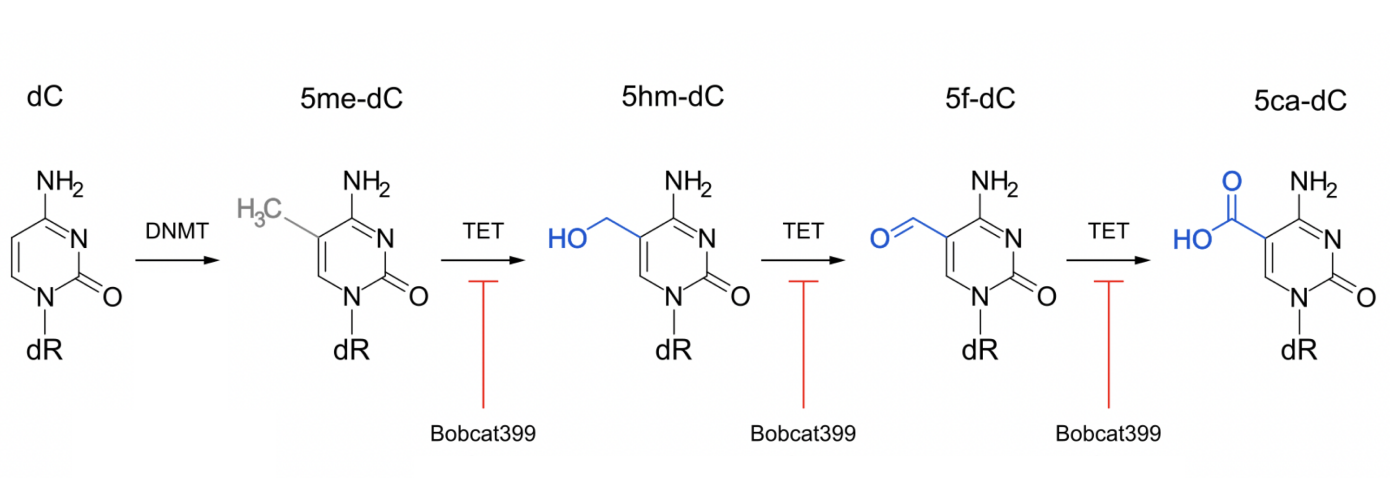
Oxidised double strand DNA also fits
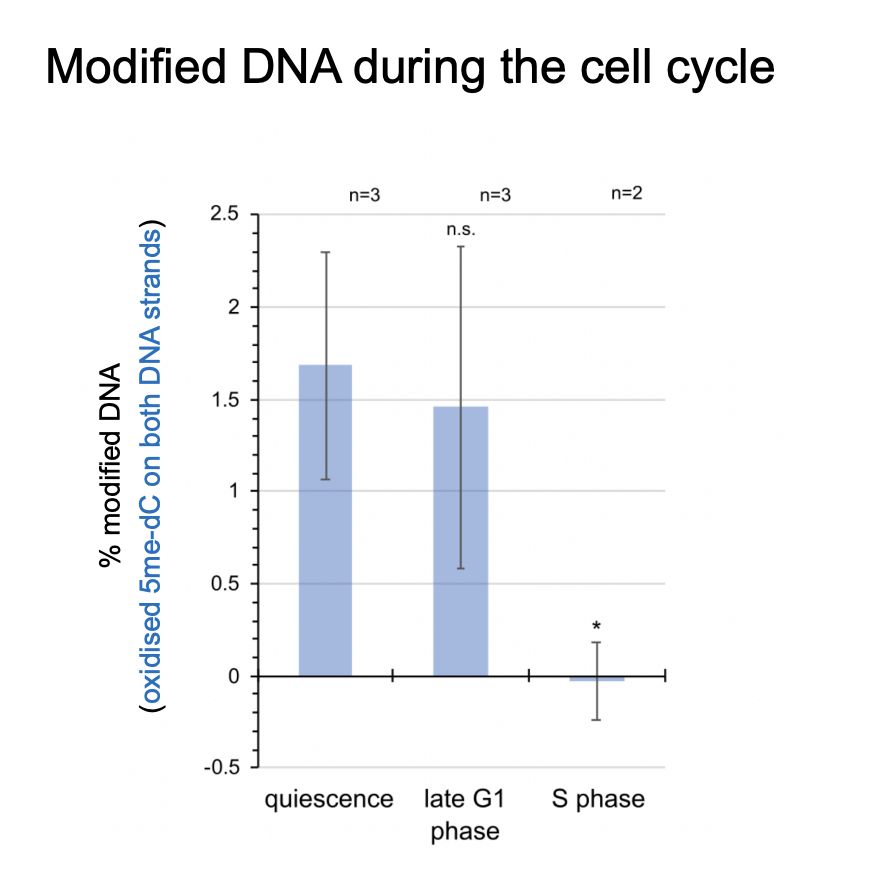
Modification of cytosine:
the oxidiased 5f-cC is only found at unreplicated origins
The oxidation reactions are inhibited by bobcat399
bobcat STOPS replication
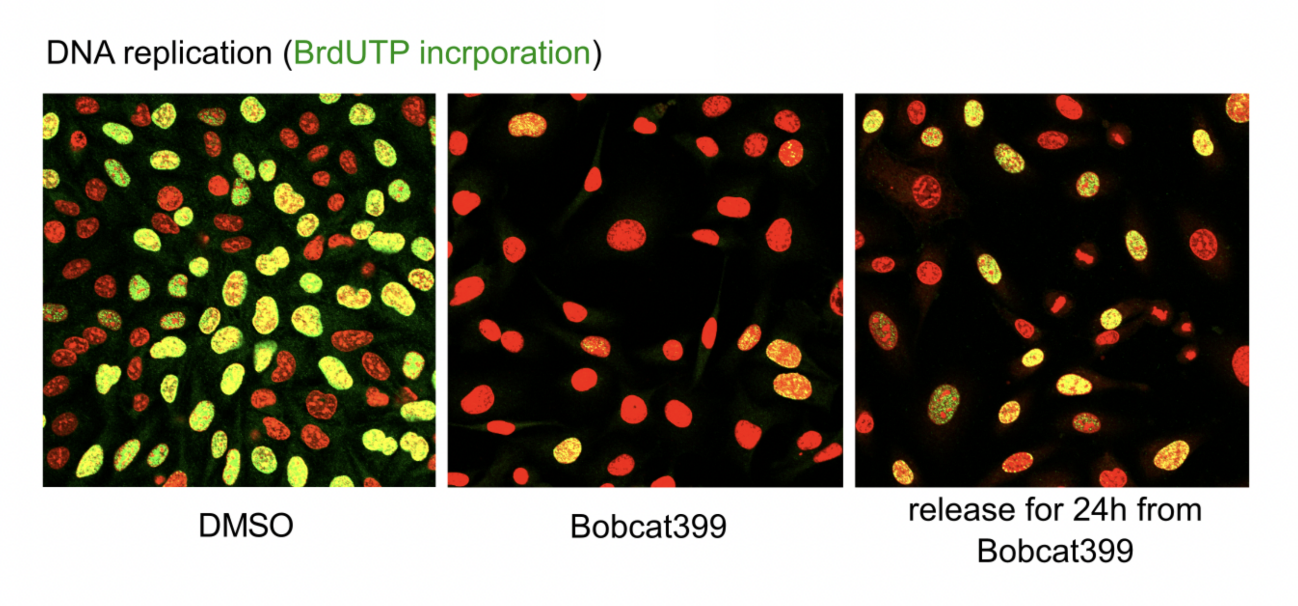
Evidence for this…
Stain with MSO→ the replicating cells will be green
Add bobcat399 enzyme→ no replicating cells (often released in S phase)
Release and add bobcat399 again→ some cells are now replicating
allows oxidation
THEREFORE: this is a cause and effeect experiement
Times when the DNA is modified
quiesnce→ after mitosis
late G1 phase→ % modified DNA still high
very low in S phase→ replicating at this point
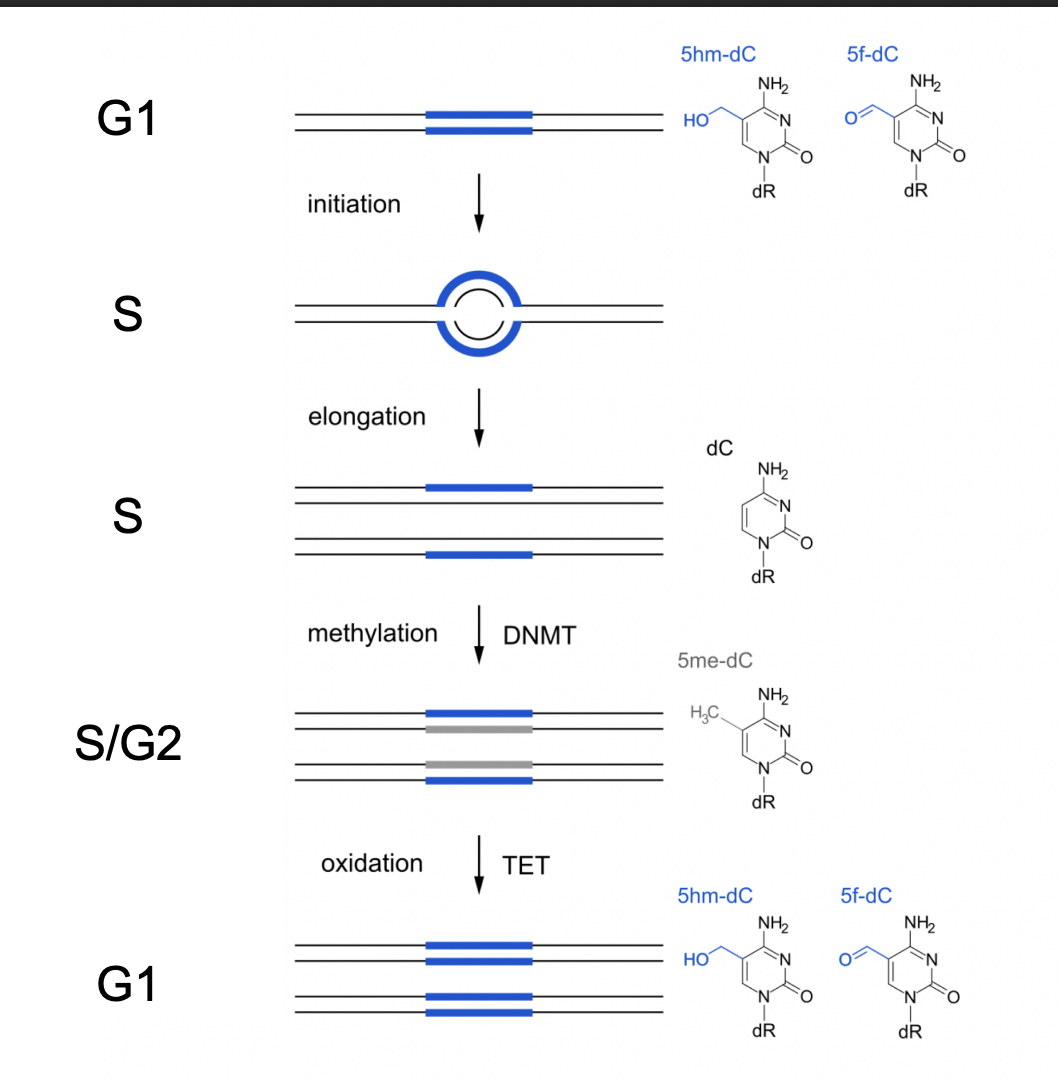
Step by step of this modification
G1→ modified so dense
initiation S phase→ the daughter strands are not dense/modified
S phase→ eongation→ some modification
S/G2→ methylation and DNMT→ becoming more dense
coming back into G1→ oxidation TET→ dense again
i.e the DNA modifications itself are helping to regulate when replication happens, to ensure that it is happening only once
Sister chromatid cohesion: newly replicated sister chromatin fibres are …
physically held together until the metaphase to anaphase transition in mitosis
COHESION
How is this sister chromatid cohesion mediated
by cohesins
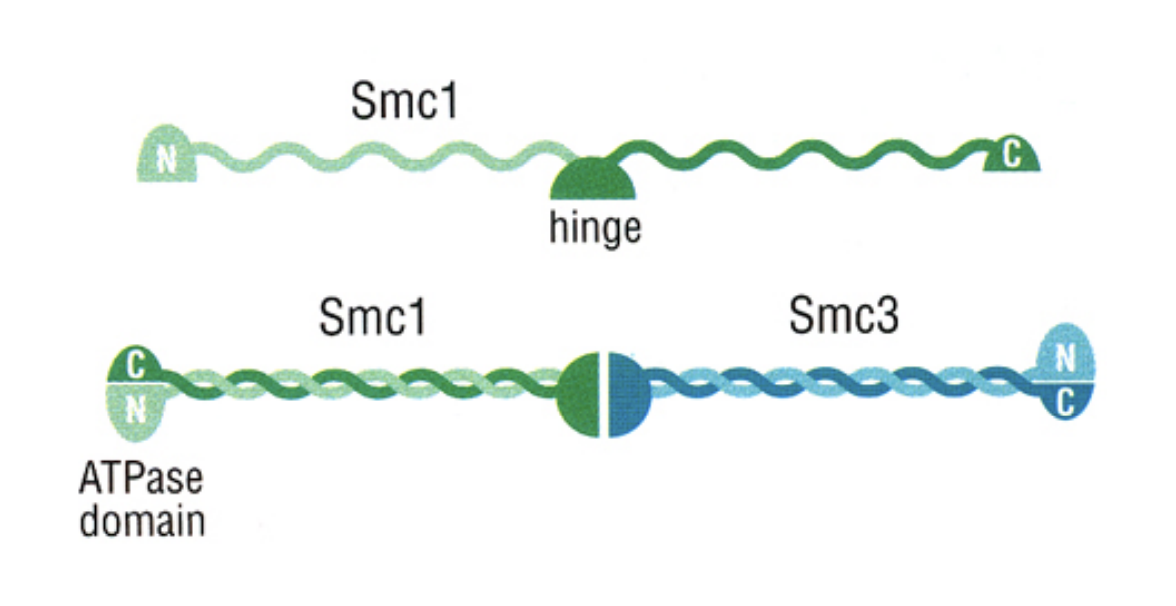
→ proteins are belonging to the class of ;structural maintenance of chromosomes’ proteins (SMCs)
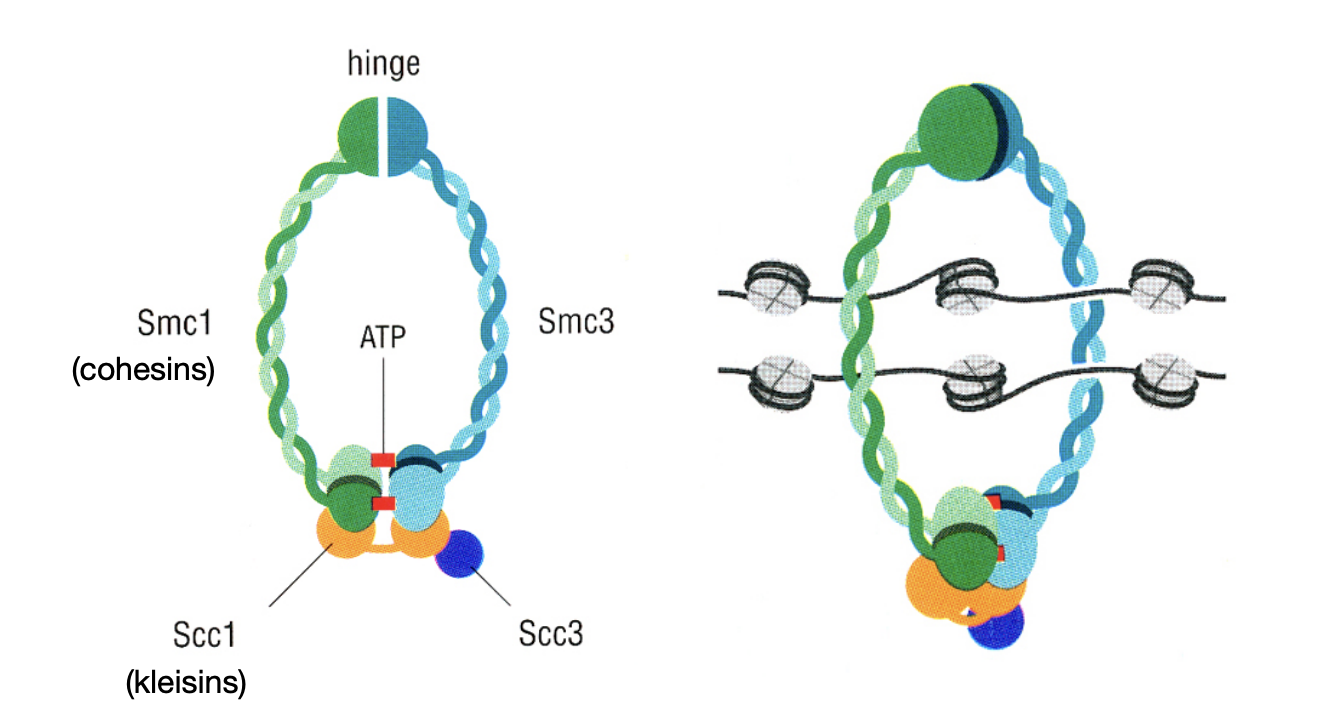
sister chromatid cohesion by SMC proteins
before they have become the kelsisin complexes
similar to cohesins seen previously but from different genes
Chromosome assembly: during chromosome replication in S phase, what must happen
entire genomic DNA must be replicated
Chromatin strucutrre also replication
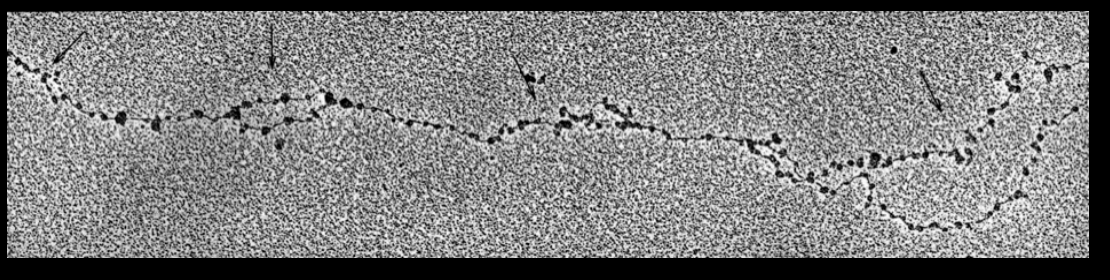
Key observations on replicating chromatin fibres:
nucleosomes are present on both unreplicated parentala and replicated daughter DNA strands
New nucleosomes are present on replicated DNA already within a few hundred base pairs past the fork
THEREFORE:
Nucleosome assembly needs to be fast and efficient
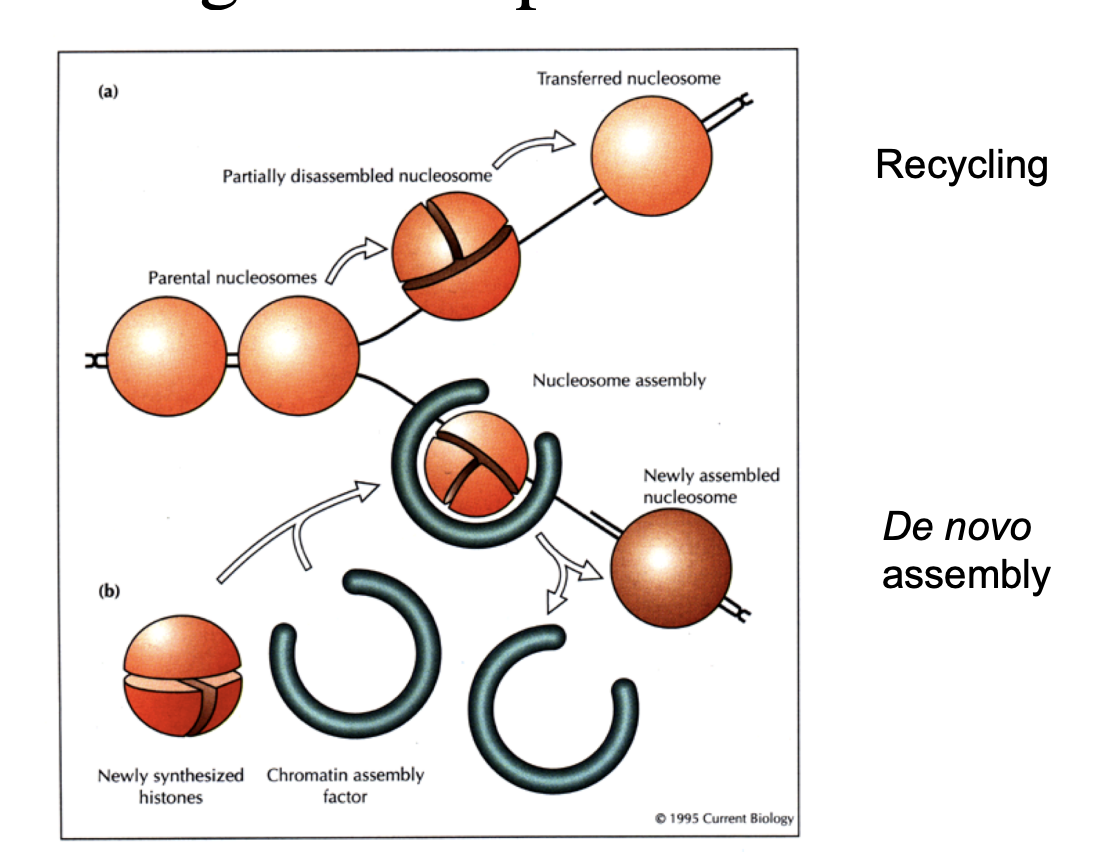
How does this happen?
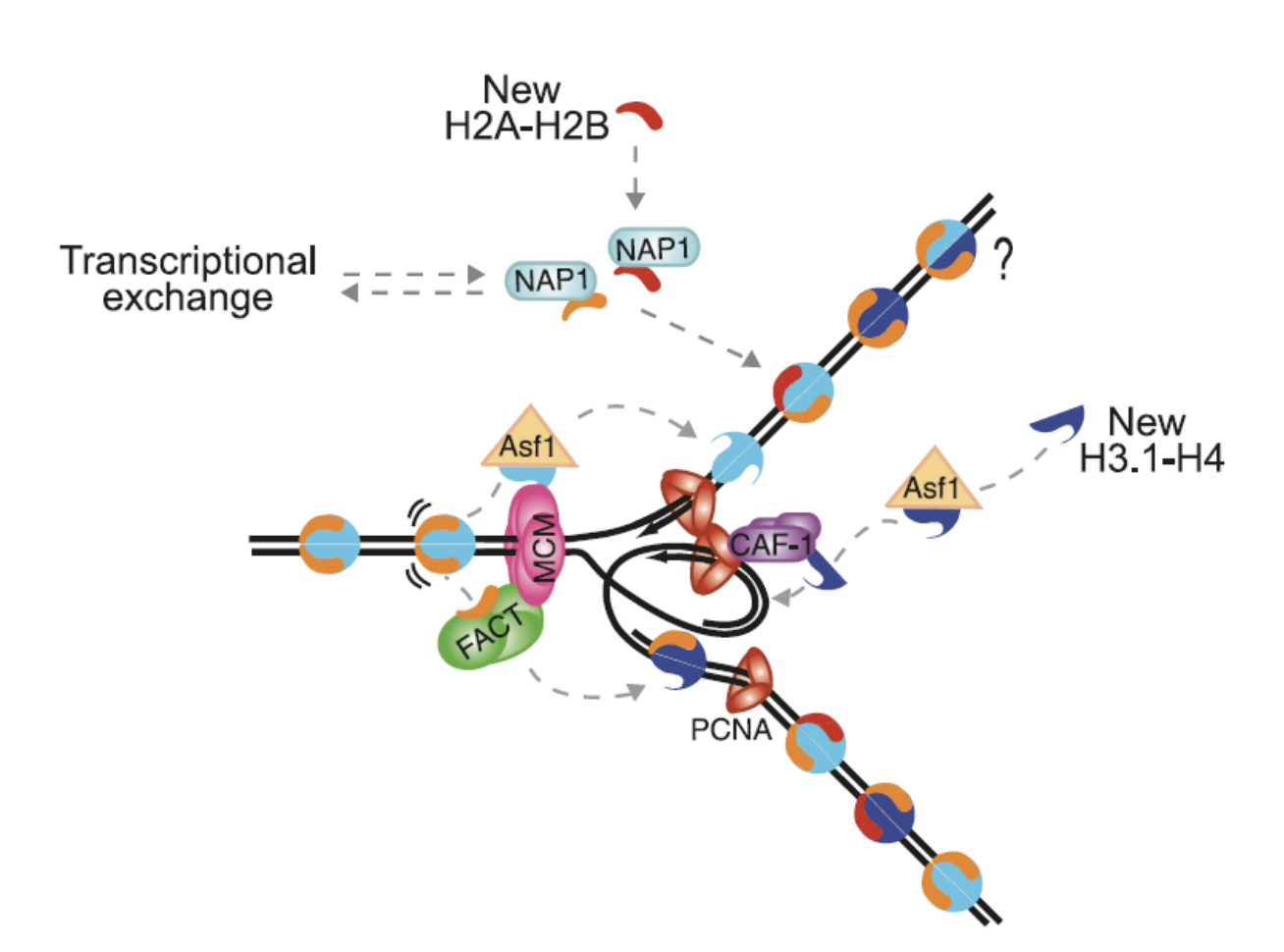
in front of advancing replication form, chromatin partially diassembles
parental nucleosoms are transfered past the replication fork machinery
their histones are recycled→ on one of the strands
new histones are synthesised during S phase of the cell cycle
assembled into nucleosomes on replicated DNA by assembly factors
In order to ensure that the histone modifications are preserved for the next DNA strand
this partial degrading helps ensure that each ‘new’ histone STILL has components of old ones
Means that the histone modifications are preserved
the cell maintains its identiity
→ still some after modification after to ensure back to normal
Issue after DNA repair?
DNA repair might mean old histone are lost or changed
so new DNA stand may not take up the whole old identity of the old histone
may lose some knowledge of expression etc of the cell type
Histones and DNA can, in principle…
self assemble to form nucleosome cores
but…
THis process is mediated by…
other proteins in the cell→ chromatin assmebly factor
Example: Xenopus embryos
proteins called N1 and nucleoplasmin
associated with histones
will assemble nucleosome cores at physiological ionic strength in vitro
Example: in human and other cells (SV40)
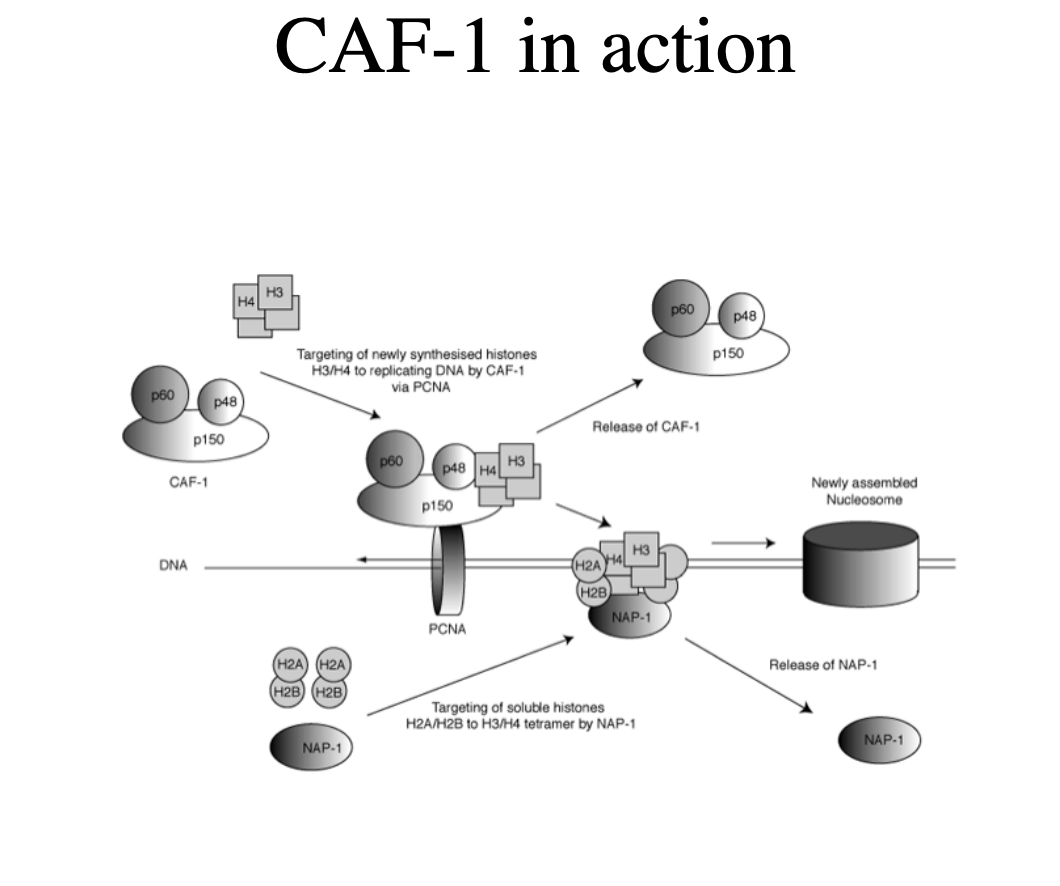
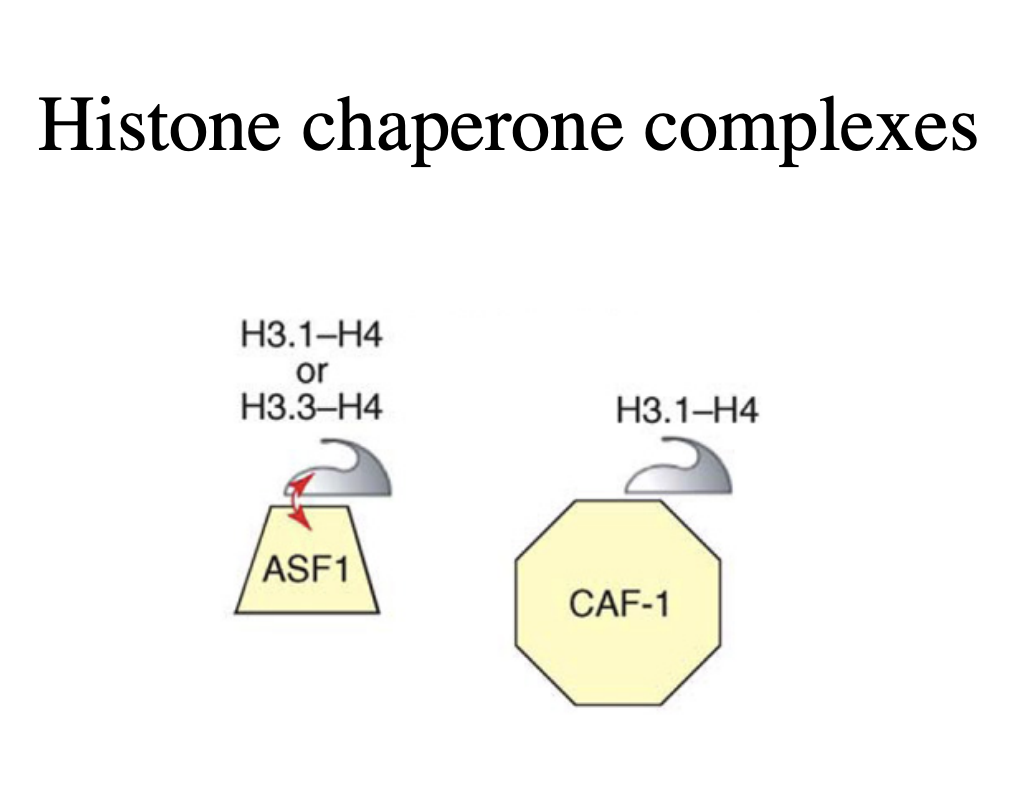
Chromatin assembly factor CAF-1 (chromatin assembly factor)
facilitiates replication-dependent nucleosome assembly in the SV40 replication system
How does it do this
interacts with replication fork protein PCNA
targets newly synthesised histones H3 and H4 to the replication fork
Other assembly proteins
Asf1 and NAP-1/2
act synergistically with CAF-1 to assemble entire new nucleosomes
OVERALL: 2 steps for histone subunits:
H3-H4 on DNA via PCNA (which was used previously in replication)
H2A-H2B dimers bind to NAP-1onto the DNA
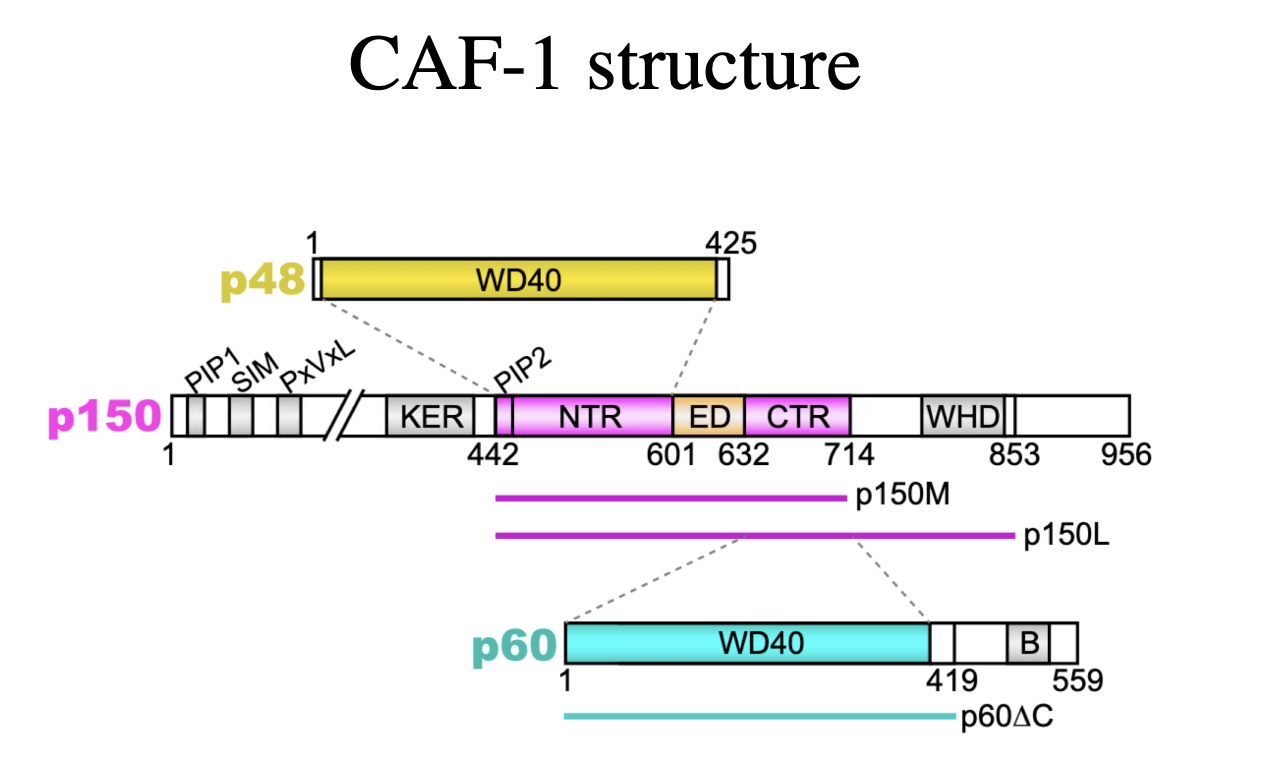
CAF-1 structure
Made up on:
p60
p48
p150
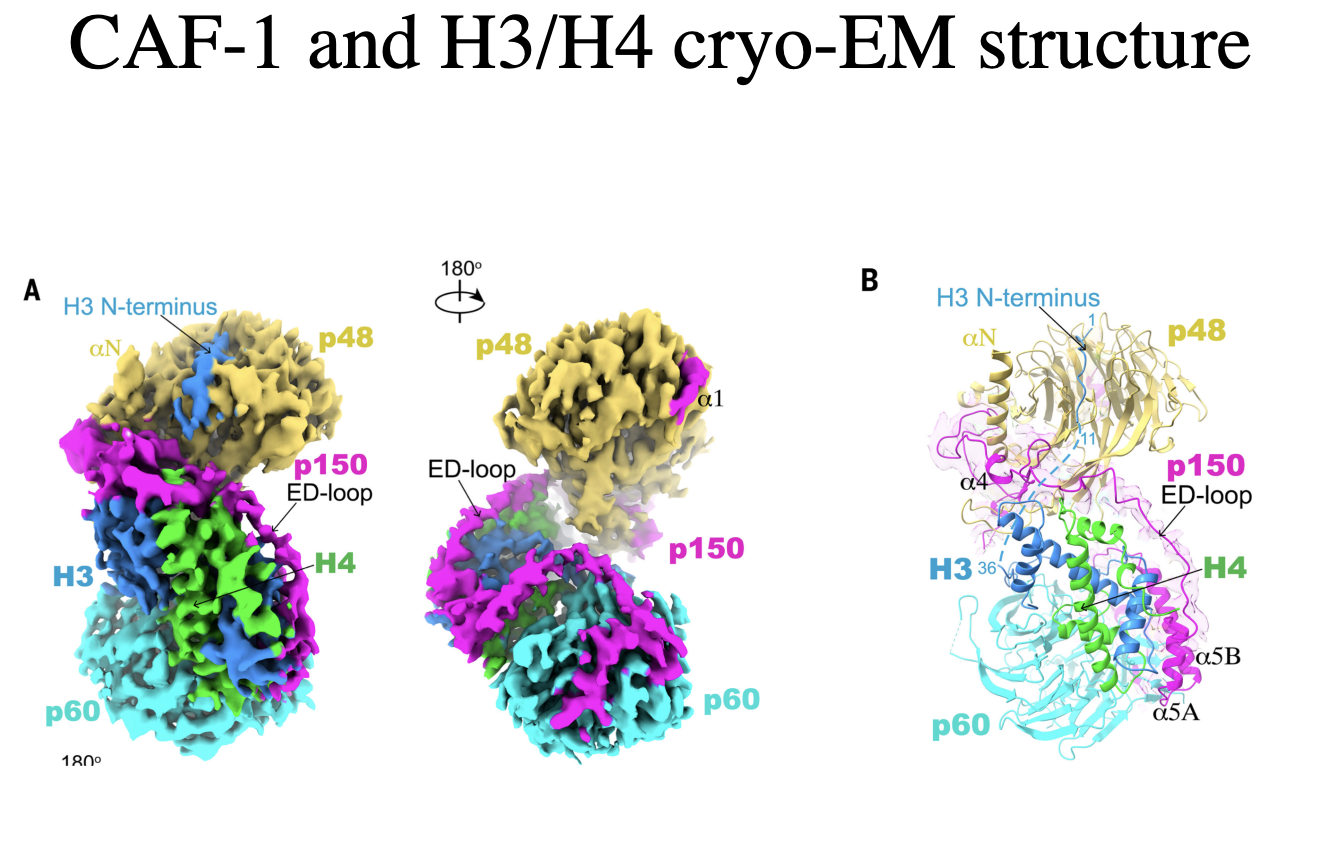
CAF-1 and H3/H4 cryo-EM strucuture
H3/H4 are in handshake arrangement
p150 loop kinda resembles the structure of DNA
in extendedn configuration
3D model of nucleosome→ histone DNA contacts
similar to the contacts being made between p150 and the histone
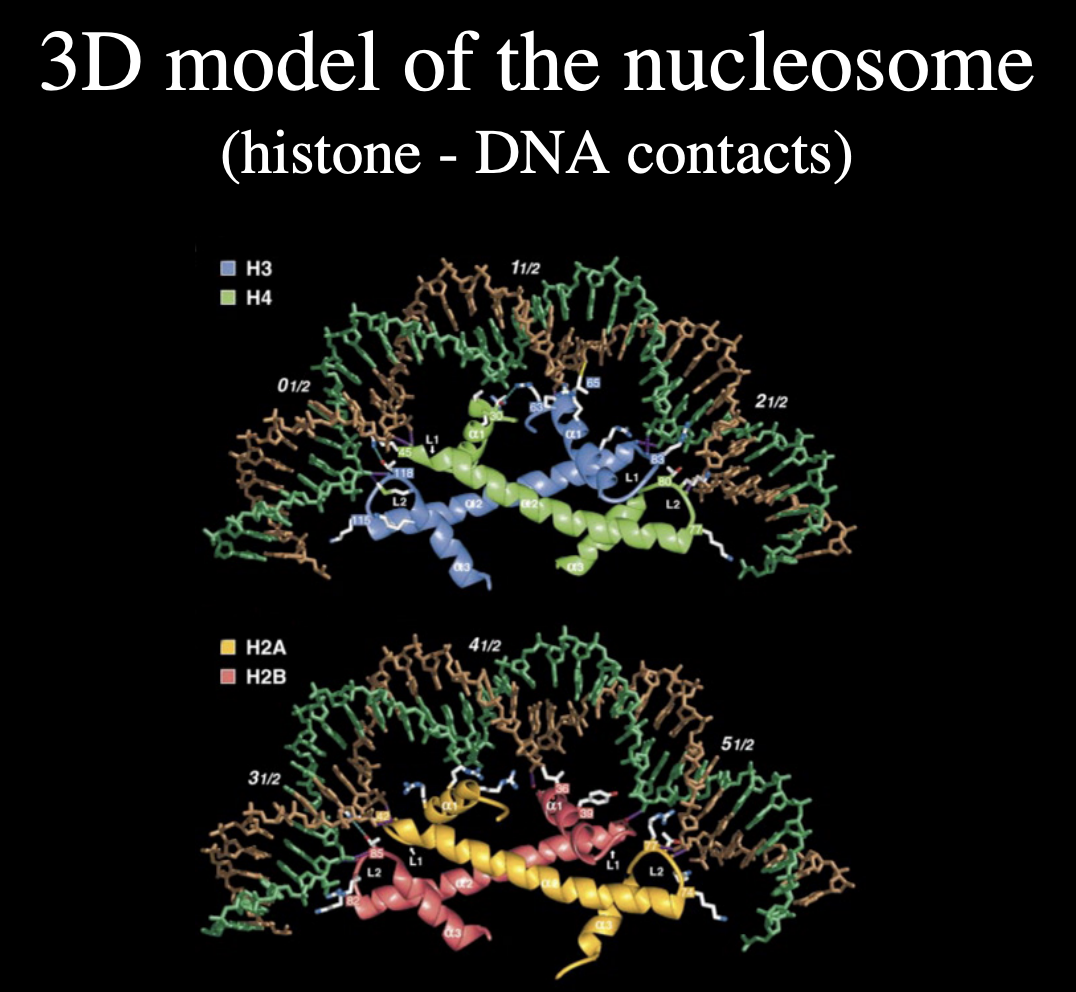
This suggests that…
CAF-1 mimics DNA
so easily hands over the histone back onto the DNA
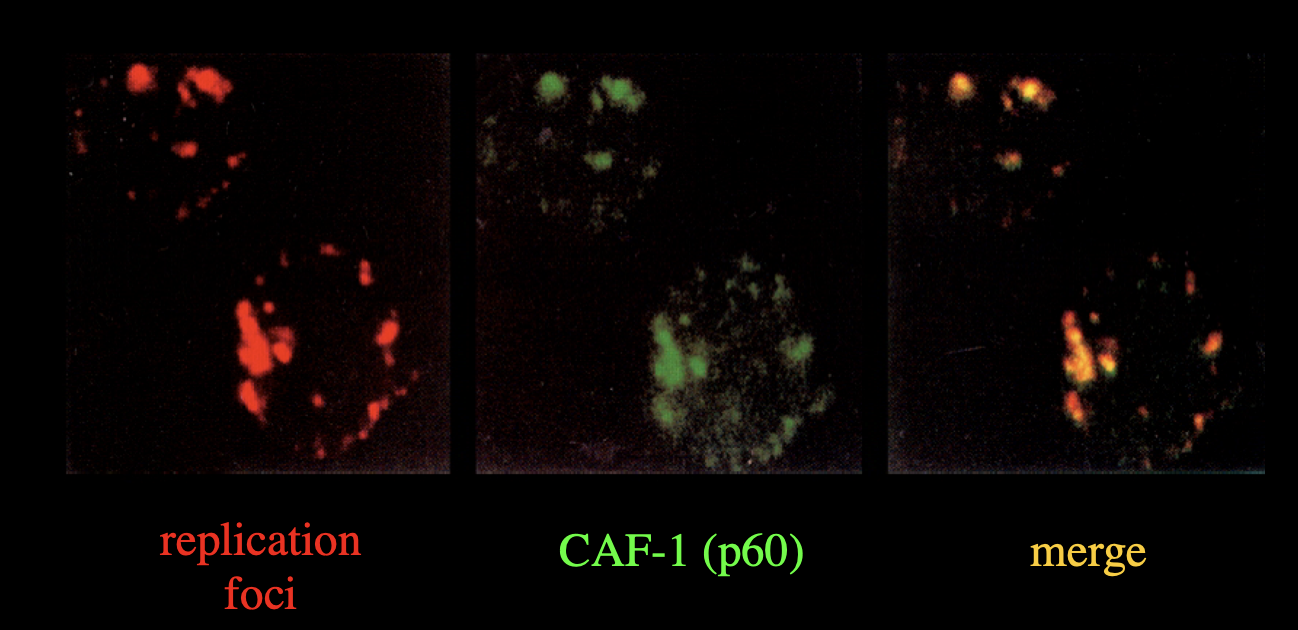
co-localises with DNA replication foci
Evidence for this
replication loci marked
CAF-1 (p60)→ shown in green
Merge→ shows orange so must be both
CAF-1 p60 colocalises with DNA replication foci
so yes this does happen in the cell
What happens to old tetramers of histones H3 and H4
stay together during replication
after transfer to a replicated DNA daughter strand
can associated with either new or old dimers of histones H2A and H2B
Linker hitones H1 associate later and higher order strucutres are forme
Chromatin remodelling
Once assembled, chromatin fibres are not static:
Factors remodel chromatin in an ATP-dependent manner
slide nucleosomes along the DNA fibre
Nucleosomes compete with DNA binding proteins
can therefore inhibit DNA trnasaction
transciption, replication initiation, repair
Types of remodelling
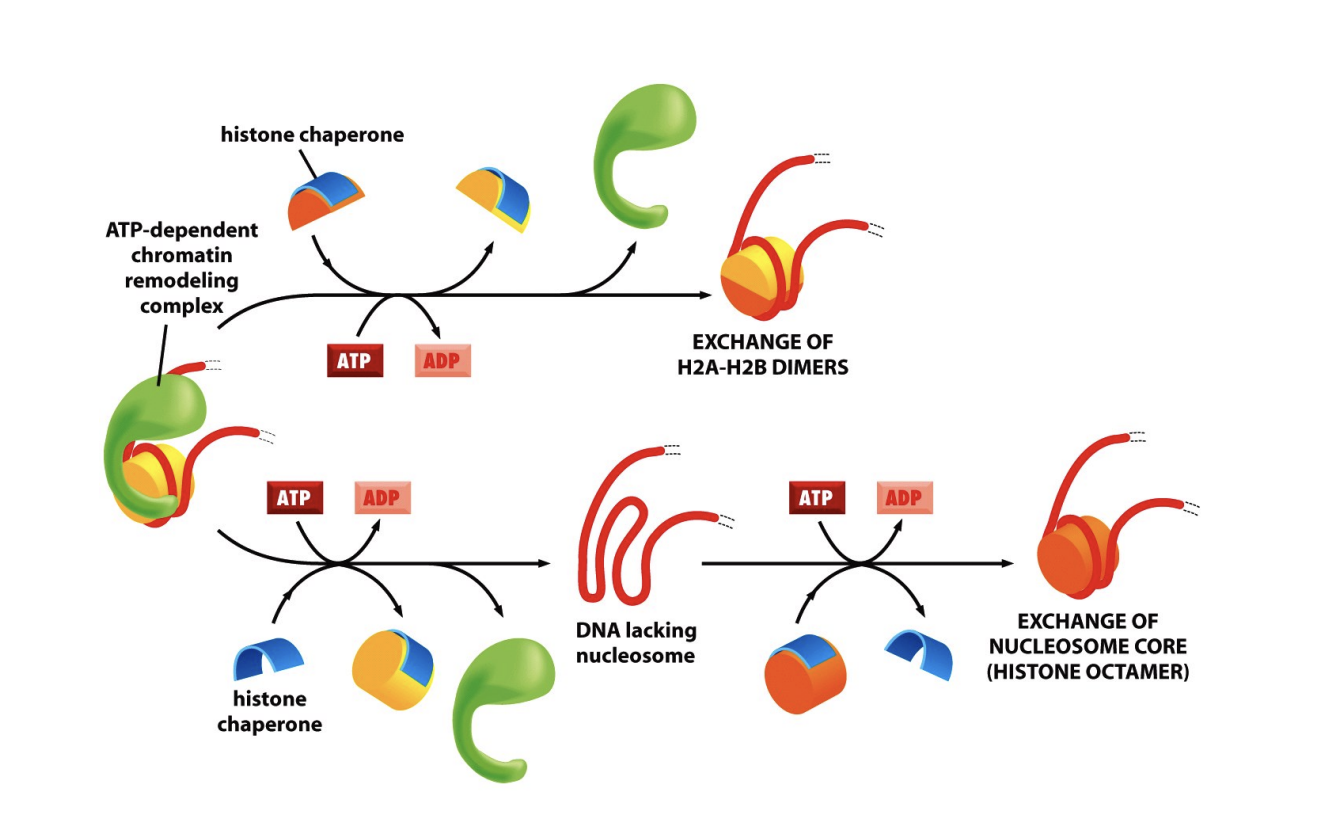
histone exchange
nucleosome sliding
These remodelling factors are usually…
large multi-subunit complexes
Histone exchange
exchange (and evict) histones within assembled nucleosomes
utilising histone chaperones as co-factors
What does this allow for?
an exchange of histone types or reprogramming of epigenetic marks
e.g histone modifications
UPPER: H2A/H2B have been switched out with a modified other
LOWER: whole histone has benn reoved and replaced with histone with new core
Nucleosome sliding
extends and makes a loop around the histone
moves around like an inchworm
gradually moving the histone along
→ takes alot of energy
each cycle moves it only by one base pair
takes energy so needs chromatin assembly factors to help
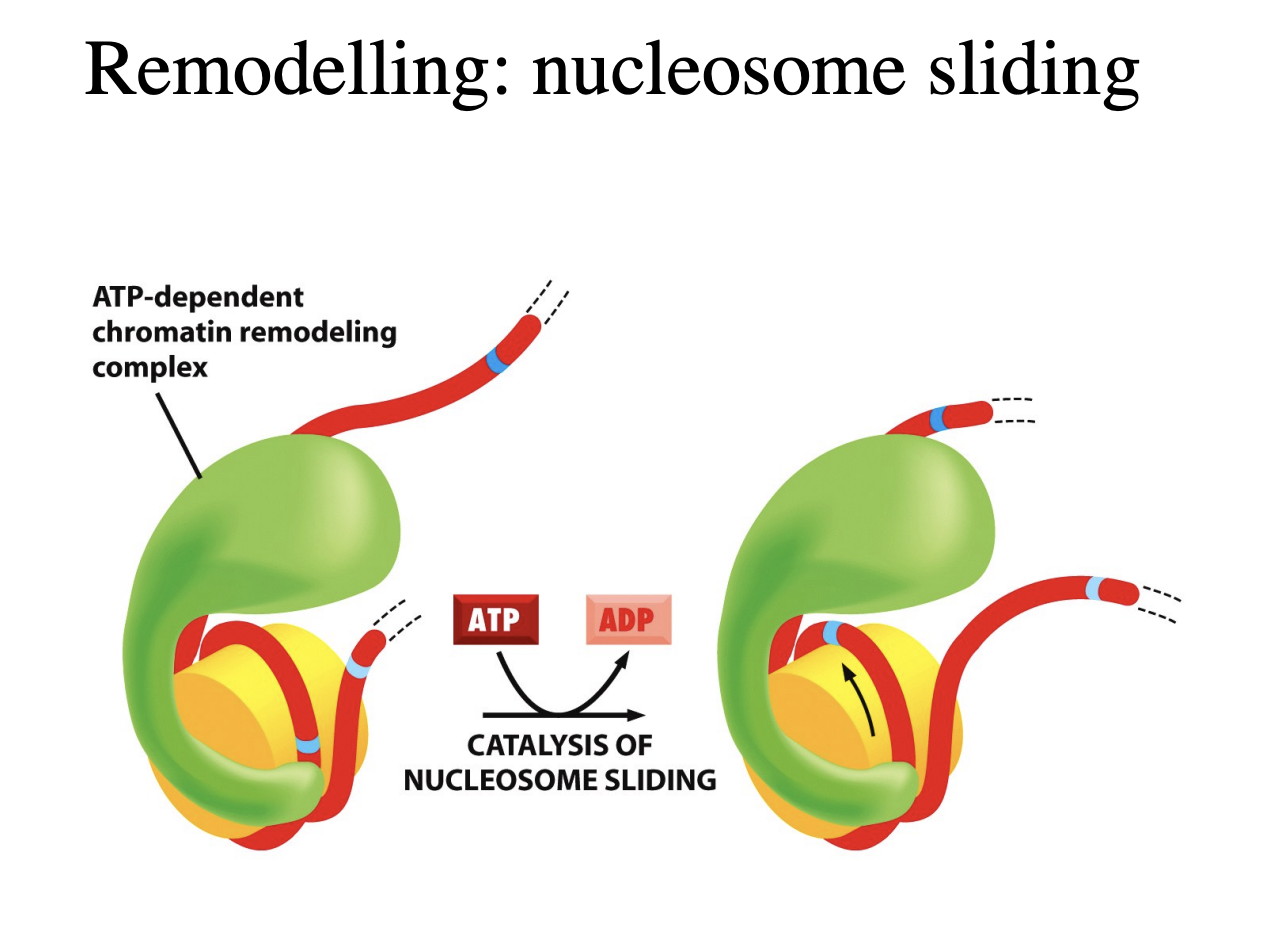
DNA translocation model (SWI/SNF)
how the nucelosome sliding works
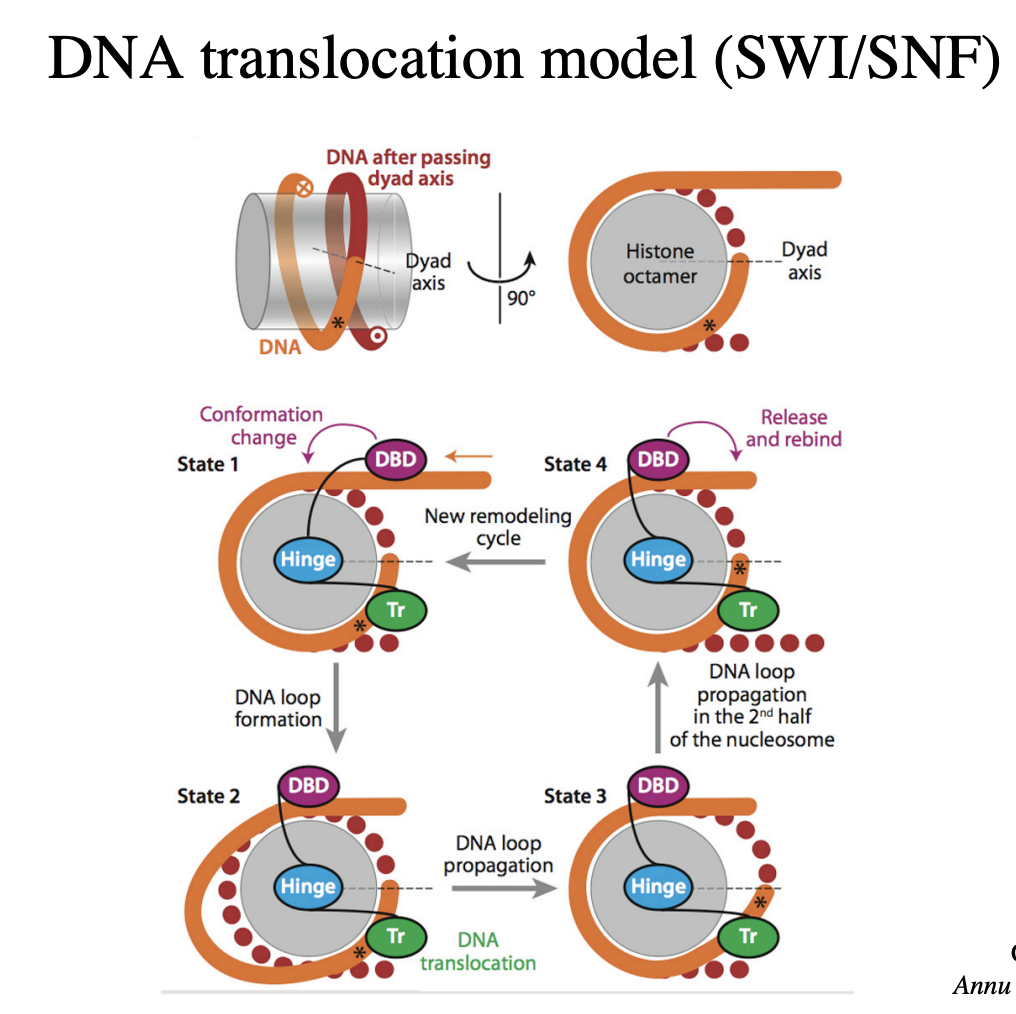
Overall: chromatin remodelling allows…
chromatin fibre to be dynamic
therefore→ can react to metabolic requirements arising from:
DNA replication, repair and transcription
A mechanistic feature of this is that…
DNA binding factors will thus be able to gain access to sites on DNA
which might be otherwise occluded by nucleosomes