Cytogenetics
1/23
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
24 Terms
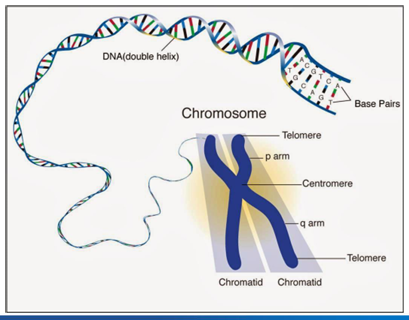
chromosome structure
-humans have 46 chromosomes
-22 pairs of autosomes and 2 sex chromosomes
-sister chromatids joined at the centromere
-ends of chromosomes capped by 6 bp repeats (TTAGGG)- telomeres
-short arm = p, long arm = q
chromosome number and karyotype nomenclature
-chromosome number described using karyotype
-karyotype: how many chromosomes are there, what sex chromosomes are present, any abnormalities
-karyotype abnormalities: extra chromosome with a “+”, missing chromosome/large piece with a “-”, translocation with lower case t and the two joined chromosomes in numerical order separated by a semicolon (t(9;22)), unidentifiable chromosome pieces called “marker” chromosomes (“+ marker chromosome” or “+mar”)
-karyotype example: (47XY+21)
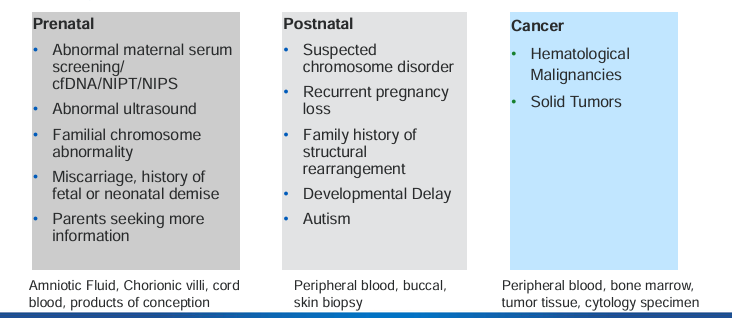
indications for chromosome and genome analysis
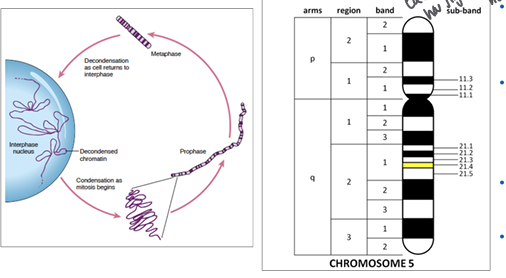
mitosis
-prophase: chromosomes condense and become visible, centrosome duplicates and migrates to opposite ends, occupies over half of mitosis
-prometaphase: nuclear membrane dissolves, chromosomes attach to the microtubules of the mitotic spindle, chromosomes begin to migrate toward equatorial plate
-metaphase: chromosomes are fully condensed and line up at the equatorial plate
-anaphase: chromosomes separate at the centromere, sister chromatids move to opposite poles
-telophase: chromosomes begin to decondense and nuclear membrane begins to reform around each of the daughter nuclei
-cytokinesis: cytoplasm cleaves
metaphase karyotyping
-cells need to be in metaphase and have the ability to divide
-keep cells in metaphase by breaking the microtubules to prevent separation
1) grow cells in culture
2) arrest cells in metaphase with colchicine (a microtubule inhibitor)
3) collect metaphase cells
4) spin in a centrifuge to pellet cells at bottom
G-banding chromosomes for visualization
-G-band staining used to identify chromosomes based on patterns of chromosome condensation
-chromosomes treated with trypsin to relax tertiary structure and stained with Giesma dye (binds nucleotide phosphate bonds)
-produces a series of dark and light bands that are unique to each chromosome
-also helps in identifying chromosomal abnormalities like deletions, duplications, and translocations
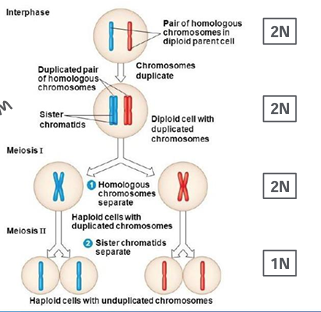
meiosis I and II
-meiosis occurs only in germ cells
-two stage cell division
-meiosis I: reductive division, separates homologous sister chromatid pairs
-meiosis II: separation of sister chromatids
-results in haploid gametes
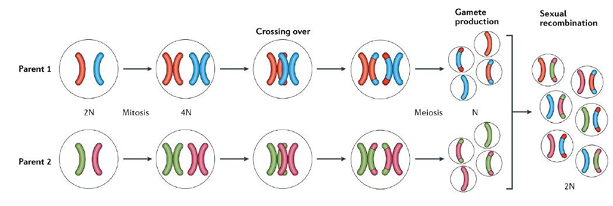
meiosis and genetic diversity
-random assortment of parental chromosomes along the equatorial plane in meiosis I
-homologous recombination (crossing over) during prophase I
sex differences in gametogenesis
-primarily lie in number of gametes produced, timing of meiotic divisions, and the cellular environment during development
-males produce a large number of sperm continuously throughout life (decreases with age), females produce a limited number of eggs during reproductive lifespan
-in females, meiosis is arrested at the first meiotic division (prophase I) until ovulation, leading to a prolonged “resting” phase (dictyate) which can increase risk of chromosomal errors
-due to prolonged meiotic arrest in females, risk of chromosomal nondisjunction (failure to separate chromosomes properly during meiosis) increases, leading to aneuploidies
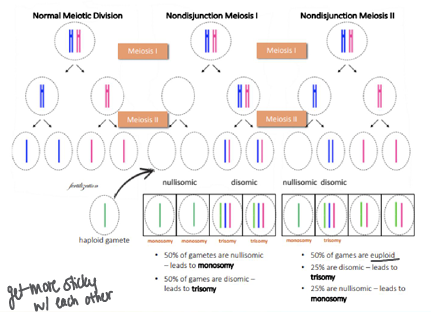
meiotic nondisjuction
-aneuploidy (unequal chromosome number) typically result of meiotic nondisjunction
-if occurs in meiosis I, maternal and paternal sister chromatid pairs fail to separate
-if occurs in meiosis II, either the maternal or paternal sister chromatids fails to separate
-monosomal gametes are not viable (except X)
-meiotic nondisjunction more commonly occurs in meiosis I
-older age is risk factor for meiotic nondisjunction → chromosomes get more sticky with each other
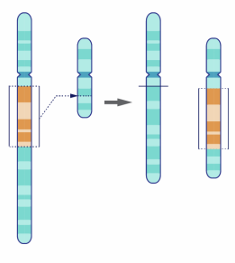
chromosomal structural abnormalities- insertions and deletions
-karyotype nomenclature: “ins(chr)”, “del(chr)”, or “dup(chr)”
-caused by errors in DNA repair
-gains/losses smaller than 5MB cannot be seen by karyotyping
-insertions: segment of chromosome inserted into another chromosome
-deletions: segment of chromosome is missing
-duplication: segment of chromosome is gain of existing chromosomal material
chromosomal abnormalities- inversions
-karyotype nomenclature: “inv(chr)”
-pericentric inversion: involves centromere and both arms of chromosome (most common cytogenetically visible rearrangement)
-paracentric inversion: involves only one arm of chromosome
-isochromosome: (karyotype nomenclature: “i(chr)”), loss of one chromosome arm, duplication of the other
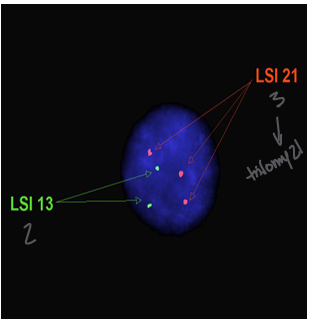
fluorescent in situ hybridization (FISH)
-use a sequence specific probe to bind DNA to ask a specific question
-probes can be designed to bind DNA or RNA and range in size 20-1500 base pairs
-hybridization: probes bind to their complementary DNA sequences on the chromosomes
-visualization: fluorescent signals visualized under a fluorescent microscope; typically 300 interphase cells are counted
combining karyotyping and FISH
-SKY (specialized karyotyping)
-metaphase spread stained with arrayed FISH probes of non-overlapping fluorescent signal
chromosomal abnormalities- translocation
-an exchange of segments between 2 chromosomes (haven’t lost any material)
-constitutional:
-prevalence: 1/500-1/1000, ~70% inherited, 30% de novo
-de novo translocations carry ~6-9% risk of major congenital abnormalitiy
-most balanced reciprocal translocations have inconspicuous phenotype; significant risk for reproductive problems including infertility, miscarriage, abnormal offspring
-if a critical gene is at the breakpoint this may influence the phenotype; submicroscopic deletion/duplications may represent cryptic non-balanced
-somatic:
-can create fusino genes
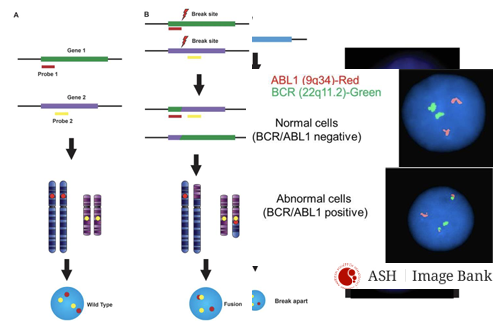
FISH for detection of translocations
-2 main techniques
-fusion probes: probes bind to regions involved in translocation; if translocation has occurred, probes will hybridize close together, producing a fused signal (yellow)
-break-apart probes: probes bind to regions flanking the translocation breakpoint; in presence of a translocation, signals will appear separated (distinct red and green)
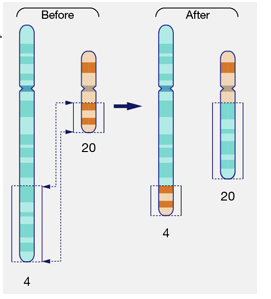
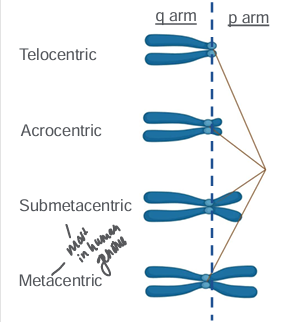
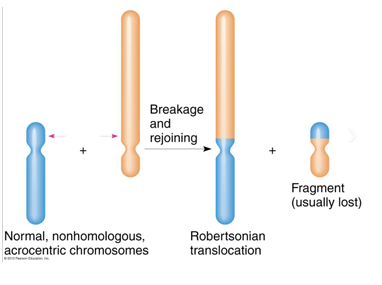
acrocentric chromosomes and rDNA redundancy
-chromosomes can be described by relative position of centromere
-no telocentric chromosomes in human genome
-p arms of metacentric and submetacentric chromosomes contain critical coding DNA that loss of a q arm would be non-viable
-**there are 5 human acrocentric chromosomes: 13, 14, 15, 21, and 22
-p arms of all 5 contain ribosomal DNA which is repetitive and redundant, which means it can be tolerated if lost
-homology between these 5 chromosomes means that they can result in translocations resulting in a single chromosome containing two q arms- called Robertsonian translocation
Robertsonian translocations
-individuals with a balanced Robertsonian translocation (carriers) have no loss or gain of significant genetic material and usually do not exhibit any clinical symptoms; have 45 chromosomes instead of the usual 46
-carriers typically have no “syndromic” phenotype, there are issues with infertility; carriers may experience increased risk of miscarriage due to production of unbalanced gametes
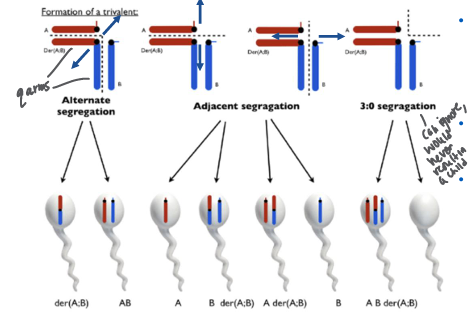
impact of Robertsonian translocations of meiosis and germ cells
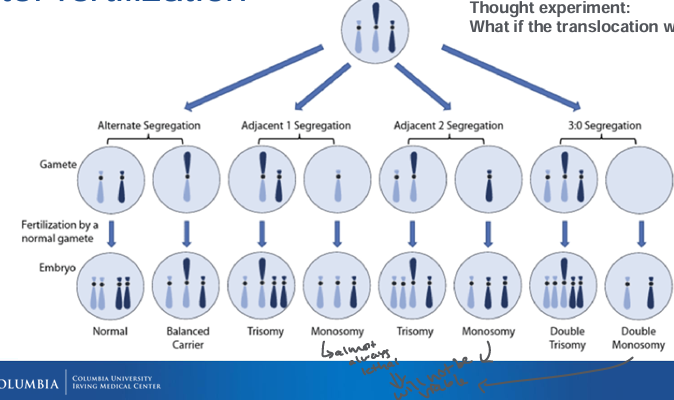
-segregation patterns: follow the centromere separation; after alignment of Robertsonian with each sister acrosomal chromosome there are 3 centromeres that need to separate
-can lead to the formation of zygotes with trisomy or monosomy
-Robertsonian translocation carriers: individuals can produce four types of gametes: normal, balanced translocation, and two types of unbalanced gametes (one with an extra chromosome and one missing a chromosome)
post-fertilization Robertsonian
chromosomal microarray
-genome-wide analysis technology used to assess DNA copy number variation (CNV) and in some cases copy-neutral alterations: loss of heterozygosity (LOH) and UPD
-first-tier test for evaluation of chromosomal imbalances associated with intellectual disability, autism, and/or multiple congenital abnormalities
-recommended for patients undergoing invasive prenatal diagnosis with fetal structural anomalies identified by ultrasound and in the evaluation of intrauterine fetal demise or stillbirth
-gives CNV info at high resolution, but can’t tell position
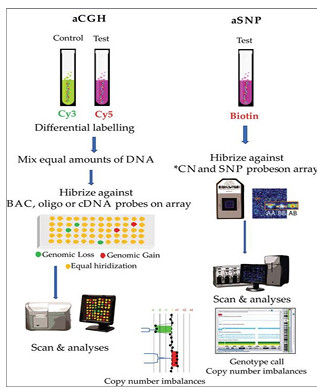
CMA steps
-DNA labeled with fluorescent dye; control sample also labeled with a different fluorescent dye
-hybridization: both DNA samples are hybridized to a microarray chip; chip contains thousands of oligonucleotide probes that correspond to specific regions of the genome
-scanning and analysis: chip scanned using a high-resolution scanner; scanner detects fluorescent signals and compares the patient’s DNA to the control DNA; comparison reveals any CNVs
-limitations: does not detect single nucleotide variants not on the SNP panel, small insertions/deletions, nucleotide repeat expansions, balanced/copy number neutral rerrangements
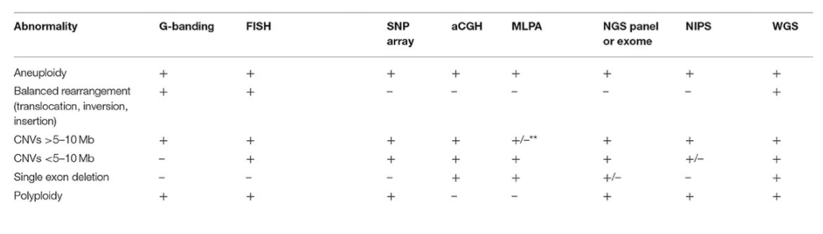
method, type of abnormalities detected, resolution, strengths, limitations- karyotype, FISH, CMA
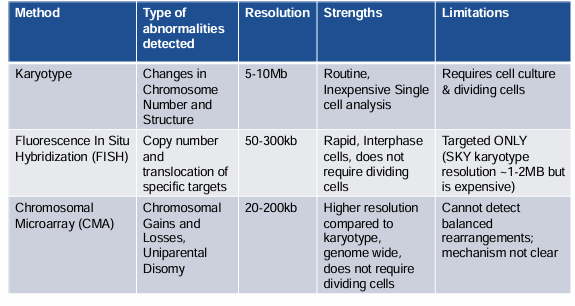
comparison of technologies available for detection of chromosomal changes and/or CNVs