MBB lecture 14-15
1/40
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
41 Terms
First step for protein purification
break open cells by sonication, shearing, and/or treatment with mild detergents
what is protein purification a first step for?
determining proteins structure and funtion
4 ways to break open cells (for purification)
sound waves pop them open
mild detergent makes holes on plasma membrane
squish chamber with piston and push cells through pore until they pop
squish cells between rotating plunger and glass vessel
how does centrifugation separate macromolecules
density (often an initial step)
what order are things moved in differential centrifugation
nuclear fraction, mitochondrial fraction, membrane fraction (then you just have cytosol fraction)
salting out
proteins=less soluble in high salt so will precipitate out
(different proteins at different salt conc.)
happen at pH where protein is neutral (net charge 0)
when are proteins least soluble
at neutral (net charge 0)
column chromatography
separates based on differential physical or chemical interactions with a solid gel matrix
types of column chromatography
ion exchange
size exclusion (gel)
affinity
ion-exchange chromatography
separates based on charge (anion exchange: protein is anionic the column is positive)
in anion exchange positive leaves first= is repelled
gel chromatography (size exclusion)
separates based on size (biggest come out first because they dont have to go in tubes)
Affinity chromatography
separates based on specific ligand interactions
(target protein interacts with ligand covalently bound to resin, non specific proteins flow through column without interacting)
SDS-PAGE: PolyAcrylamide Gel Electrophoresis
separates based on mass
preformed on vertical apparatus, buffers in top and bottom chambers, 2 chambers connected by gel matrix between 2 glass plates, sample added to top with pipette, external power source allows current to flow through, migration of negatively charged proteins toward anode (positive) side of chamber
Estimation of protein MW in SDS-PAGE: use MW markers
The electrophoretic mobility of proteins in SDS-polyacrylamide gels is inversely proportional to the log of their MW
what is a dye front
leading edge of dye migration during electrophoresis Rf: retention factor
Rf=
distance migrated by protein divided by distance migrated by the dye front
protein ladder
mixture of proteins of known molecular weights that run alongside unknown protein samples during electrophoresis process. A reference to estimate molecular weights
2D PAGE
used to separate proteins based on both pl and MW.
separated in first dimension by isoelectric focusing (based on overall charge). then separated in 2nd dimension by standard SDS-PAGE
protein spots can be excised from gel and identifies using mass spec.
what is 2D DIGE
2-d differential in-gel electrophoresis
what dyes does 2d DIGE use and for what
Cy3 (green) and Cy5 (red) to distinguish 2 protein samples run on the same 2D PAGE gel
(proteins unique to one sample show one colour, proteins in both samples have a mixture colour)
what is proteomics
large-scale study of proteins (mostly structure and function)
does proteomics usually refer to the entire set of proteins for an organism
yes usually refers to that (proteome) or set of proteins present in particular cell type under certain conditions
does the proteome differ from cell to cell and change?
yes and constantly changes depending on interactions with environment
tools for proteomics
isoelectric focusing, protein arrays and chips, co-immunoprecipitation (co-IP), mass spec (MS)
approaches are, by necessity, “high-throughput” meaning lots of proteins studied/processed at the same time
the proteomics approach (steps) (7)
identifying all proteins in given sample
sample (could be for ex. lysate from organ, secreted proteins from bacterium, cytoplasmic contents, membrane fraction)
prep steps
isolate proteins (1D PAGE, 2D PAGE)
cut protein spots out of gel
digest (cut up) with protease (trypsin)
mass spec
protein id and informatics
NMR (6 points)
nuclei align in electromagnetic (em) field according to spin
nuclei can “flip” if given pulse of em radiation at “resonance frequency “
energy absorbed to flip nuclei is recorded as line or peak on an NMR spectrum
energy required for flip depends on surroundings of nuclei
spectrum gives info about structure
proteins need to be little (<50kDa)
X-ray crystallography (7 points)
expose protein crystals to x-ray beam, collect x-ray diffraction data
big things like viruses can use this method
need to be able to crystallize protein (getting diffraction quality crystal is rate limiting step)
crystals are grown in buffer containing “precipitant” that reduces solubility of protein and favours formation of ordered crystals
in protein crystal all molecules are oriented the same way in a 3D lattice
proteins not fully hydrated so not in native form/environment
membrane proteins need detergents to solubilize them (hard to crystallize)
what are antibodies
also called immunoglobins are proteins made by animals in large amounts response to presence of antigen
what are antigens made of
protein, polysaccarides, nucleic acids, small molecules
why are antibodies useful
because of high specificity
they bind to their antigen with high specificity and affinity
can be used to identify proteins and their locations in the cell, to purify proteins, quantify them, and to identify protein binding partners
antibody structure
made of 4 chains (2 heavy, 2 light)
each heavy and light pair form 2 identical antigen-binding sites
chains are held together by disulphide bonds and non-covalent interactions
what is the F(ab) domain (antibody) and what site on the antigen does it bind to
antigen-binding fragment, epitope
what is the F(c ) domain (antibody)
binds to receptors on host cells (crystallizable fragment)
why do antibodies bind to antigens
stereochemical complementarity between anitgen binding site and epitope on the antigen
antibody production (rabbit example) (8 steps)
immunize 4 times over 6 weeks
obtain blood sample
blood serum contains mixture of nonspecific and antigen specific antibodies
purify using affinity chromatography (load, wash, elute)
after load flow through fraction comes through
after wash nonspecific antibodies come through
after elute elution of antigen specific antibodies
end up with affinity-purified rabbit polyclonal antibodies
purpose and steps for Enzyma linked immunosorbent assay (ELISA) (7 steps)
to quantify the protein antigen based on amount of primary Ab bound
antigen coated well
wash
specific antibody binds to antigen
wash
enzyme linked antibody binds to specific antibody
wash
substrate added and converted by enzyme into coloured product (rate of colour formation is proportional to amount of anitbody)
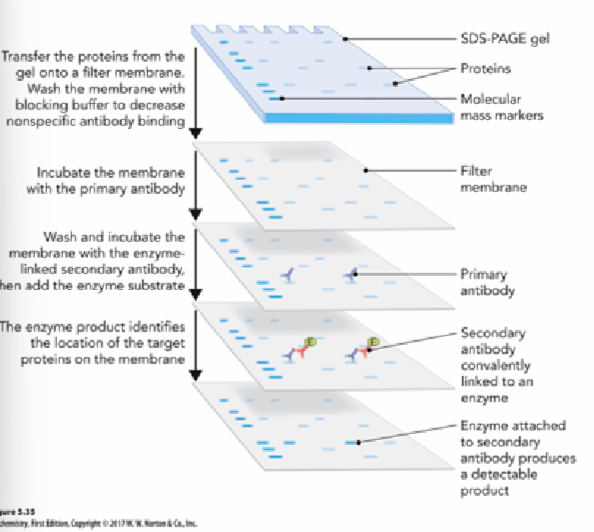
what is immunoblotting (western blots)
antibody based detection and quantification of protein antigen of interest.
protein has been electrophoresed on SDS-PAGE gel and transferred to membrane prior to incubation with primary antibody
steps of immunoblotting
what is immunoflourescence
an antibody-based technique used to identify proteins in cells that have been chemically treated in a way that preserves cell architecture