Proteins and enzymes
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
22 Terms
probability of going to S to P at the top of the energy barrier where the reaction intermediate is?
equal probability, thats why enzymes rearrange bonds and change position of chemical groups to push the reaction towards products
methyl group
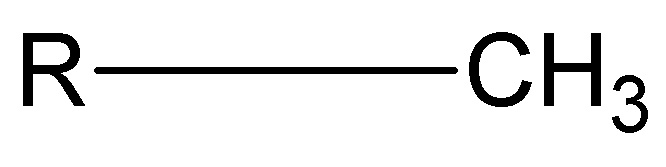
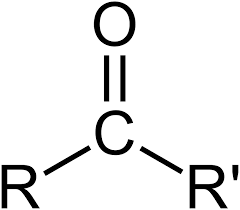
carbonyl group
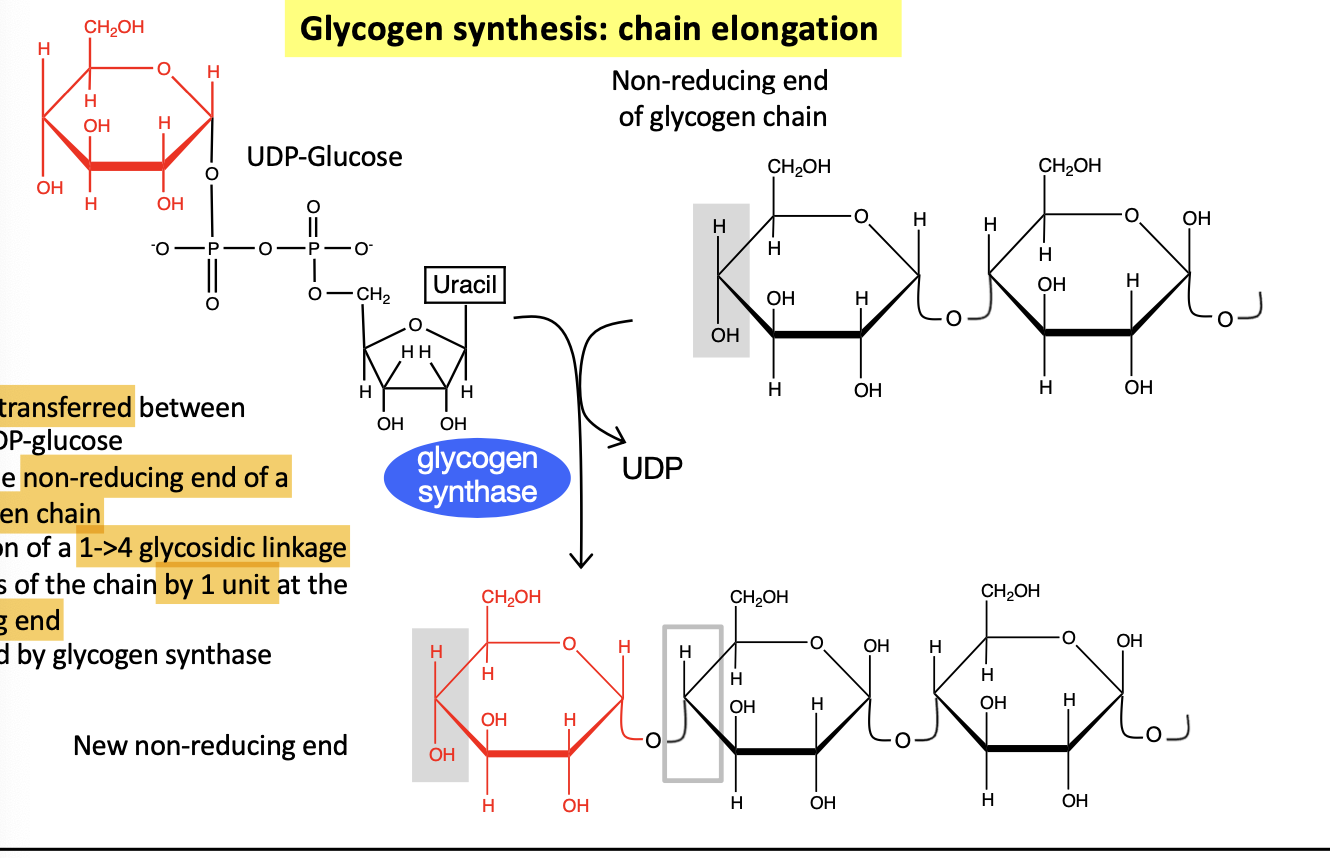
glycogen chain synthesis steps
Use ATP to phosphorylate glucose on C-6
catalyzed by hexokinase in muscle and glucokinase in the liver
Move phosphate from C6 to C1 —> glucose 6-P to glucose 1-P
React with UTP to form UDP-glucose and 2 phosphates
this activates it
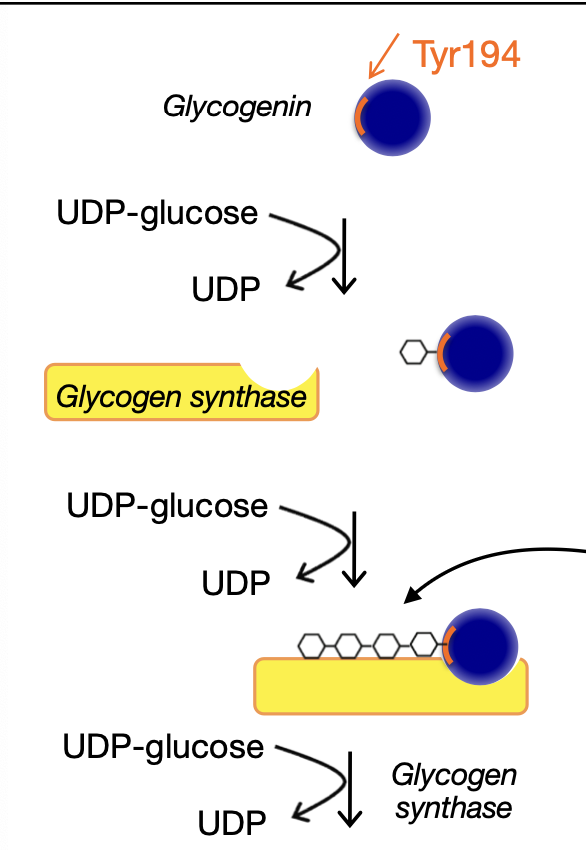
Glycogenin is an enzyme that has a tyrosine residue and UDP-glucose attaches to the residue
Glycogen synthase is recruited and glycogenic fits attaches to it
PP1 binds to R5 which activates it which then the PP1-R5 complex dephosphorylates GS which activates GS for chain elongation
Glycogenin adds another UDP-glucose at the C4 non reducing end to produce UDP and a 1-4 linkage which adds to the non-reducing end of a chain
Glycogen synthase takes over the elongation by adding more UDP-glucose to form a long chain
Branches get added by glycogen synthase and branching enzyme working together to form a highly branched chain
Regulation inactivates glycogen synthase and releases it from glycogenin but the reducing end of glycogen stays connected to glycogenin
branching enzyme function
cleaves 1-4 linkage in a glycogen chain and attaches broken chain to another chain to form a 1-6 bond to form a branch
glycolysis steps
Glucose converted to glucose-6-phosphate by hexokinase
uses 1 ATP
irreversible
G6P converted to fructose-6P (furanose) by phosphoglucose isomerase
F6P converted to Fructose 1-6-biphosphate by phosphofructokinase
uses 1 ATP
irreversible
F1-6BP converted into DHAP and glyceraldehyde 3 phosphate by aldolase
DHAP and G3P in equilibrium but DHAP is converted into G3P by triose phosphate isomerase
G3P converted into 1,3 biphosphoglycerate by glyceraldehyde 3-phosphate dehydrogenase x2
reduces NAD+ to NADH (exergonic)
uses P to phosphorylate (endergonic)
1,3 BPG converted to 3 phosphoglycerate by phosphoglycerate kinase x2
high energy —> converts ADP to ATP
3PG converted to 2PG by phosphoglycerate mutase x2
2PG converted to phosphoenolpyruvate by enolase x2
hydrolyses it so H2O is formed in the process
PEP converted to pyruvate by pyruvate kinase x2
super high energy —> converts ADP to ATP
irreversible
bypass steps of gluconeogenesis (converting pyruvate to PEP)
First bypass
Pyruvate transported from cytosol to mitochondria and alanine (product of pyruvate) transported into mitochondria and then converted to pyruvate
Pyruvate converted into oxaloacetate by pyruvate carboxylase
uses 1 ATP
uses CO3- as source of carbon
Oxaloacetate converted into malate by malate dehydrogenase
uses oxidation of NADH to NAD+
Malate exported to cytosol where it is converted to oxaloacetate
reduces NAD+ to NADH
Oxaolacetate converted to PEP by PEP carboxykinase
Uses 1 GTP
In cells with lactate after fermentation (muscle and red blood cell)
Lactate converted to pyruvate in cytosol
by lactate dehydrogenase
NAD+ to NADH
Pyruvate goes to mitochondria where it gets converted to oxaloacetate
pyruvate carboxylase
1 ATP, CO3-
Oxaloacetate converted into PEP
mitochondrial PEP carboxykinase
PEP transported out of cell into cytoplasm
Second bypass (one step)
Fructose 16BP converted to F6P
F16biphosphatase
exergonic, but not coupled with ATP synthesis
Coupled with regulation of PFK1 which is the exergonic reverse bc u cant have both exergonic reverse reactions running at the same time in the cytosol
Third bypass (one step)
Glucose6P to glucose
glucose-6-phosphatase which is only found in liver and kidneys
Exergonic and irreversible but step in glycolysis is more exergonic
pyruvate dehydrogenase steps
decarboxylation
E1 which has TPP attached to it and it reacts with pyruvate forming CO2 and hydroxyethyl TPP
Oxidation
Lipoamide arm bound to E2 goes into E1 where the transfer of acetyl group from TPP to lipoamide arm
thioester (big energy of hydrolysis) between lipoamide and acetyl group is formed and TPP back in its original state
Tranfer
arm goes into second E2 unit and the thioester bond is broken but a new one is formed between acetyl group and coenzyme A to form acetyl-CoA
Regeneration
The disulfide bond on the arm has to be regenerated again so it goes into E3 which has FAD which will accept 2 electrons and 2 protons released when disulfide bond is formed to form FADH2
FAD needs to be regenerated so give protons to NAD+ to form NADH which will be used in oxidative phosphorylation
Citric Acid cycle step 1
Acetyl-CoA + Oxaloacetate to Citrate
Citrate Synthase
Uses H2O and produces CoA-SH
Irreversible
Cycle Step 2A and B
A: Citrate to cis-Aconitate
B: cis-Aconitate to Isocitrate
Aconitase
removes an OH and H to form H2O in 2A, adds in water in 2B to reposition OH
reversible
Cycle Step 3
Isocitrate to a-Ketoglutarate
Isocitrate Dehydrogenase
Forms CO2 and NADH
irreversible
Cycle Step 4
a-Ketoglutarate to Succinyl Co-A
a-Ketoglutarate dehydrogenase complex
reacts with CoA-SH to form CO2 and also formed NADH
irreversible
Cycle Step 5
Succinyl Co-A to Succinate
Succinyl Co-A synthetase
GDP to GTP, and CoA-SH produced
reversible
high energy
Cycle Step 6
Succinate to Fumarate
succinate dehydrogenase
Produces FADH2 and removes and H
Cycle Step 7
Fumarate to Malate
fumarase
uses water to add an OH group
reversible
Cycle Step 8
Malate to Oxaloacetate
malate dehydrogenase
removed 2 H and produced NADH
reversible
Complex 1
NADH-ubiquinone oxidoreductase
FMN accepts 2e and 2H+ from NADH + H in the matrix
FMNH2 gives 2e to Fe-S
Fe-S gives 2e to Q and matrix gives 2H+ to Q
QH2 is formed
4 H+ get pushed into intermembrane space
Complex II
Succinate dehydrogenase
this protein in step 6 of the TCA cycle produced FADH2
Then it gives 2e- to Fe-S
gives 2e- to Q
forms Q2
Complex III
Q cytochrome c oxidoreductase
gets 2e- from QH2
Gives 4H+ into the inter membrane space
Through the Q cycle 2 cytochrome Cs each shuttle 1 electron to Complex IV
Q cycle
Q has 3 states
Ubiquinone
Q
Radical Semiquinone → gains an e- and p
QH
Ubiquinol → 2e- and 2p (fully reduced)
QH2
QH2 from Complex I and Complex II goes to Complex III to become Q again but cytochrome C in Complex III can only accept 1 e- at a time
How it works
Has 2 binding sites: Q0 and Qi
QH2 binds to Q0 and a Q binds to Qi
2 H+ protons from QH2 get sent into intermembrane space
Has 2e- left
One e- is transported to Fe-S, then cyt C1, then cyt C to accept 1e- which is on the intermembrane side of Complex III
Cyt C dissociates from Complex III
Other e- from QH2 is given to cyt BL, cyt BH, and then to Q at site Qi
This forms a Qe- at the Qi site and a Q at the Q0 site
The Qe- takes one H+ from the matrix to form QH
The Q at the Q0 site is replaced by another QH2 and cytochrome C is replaced
The QH2 releases its 2H+ into intermembrane space
1e- is sent to cyt C
The other e- is given to QH at Qi site and it also takes another H+ from matrix to form a QH2 at the Qi site
Qi releases QH2 and Q0 releases Q
Complex III can start a new cycle
So one round in total releases 4H+ into intermembrane and takes 2 H+ from matrix
Complex IV
cytochrome oxidase
Step 1
takes 1 e- from the first cyt C
gives it to Cu-A, then Heme A, then Heme A3, then Cu-B
Step 2
takes 1 e- from second cyt C
gives it to Cu-A, then Heme A, then Heme A3
O2 is put in between reduced Heme A3 (which is now Fe2+) and reduced Cu-B (which is now Cu+)
Forms a bridge: Fe2+-O2-Cu+
Step 3
takes an e- from another cyt C
gives it to Cu-A, then Heme A, then bridge
One H+ is extracted from matrix
H+ breaks bridge by formed an OH
Left with: Fe2+-O and OH-Cu+
Step 4
takes e- from another cyt C
gives it to Cu-A, then Heme A, then Fe2+-O
take another H+ from matrix
Form: Fe2+-OH and OH-Cu+
Step 5
take 2 more H+ from matrix to form 2 H2O molecules from the 2 OH groups
Left with: H2O, Fe3+, and Cu2+
Shuttles 4 protons into inter membrane space (total of 8 protons lost from matrix), 2 protons per molecule of H2O