BIOL215 (copy minus early price stuff)
1/488
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
489 Terms
Autotrophs
Obtain carbon from inorganic sources (CO2), self feeder like plants, phytoplankton etc
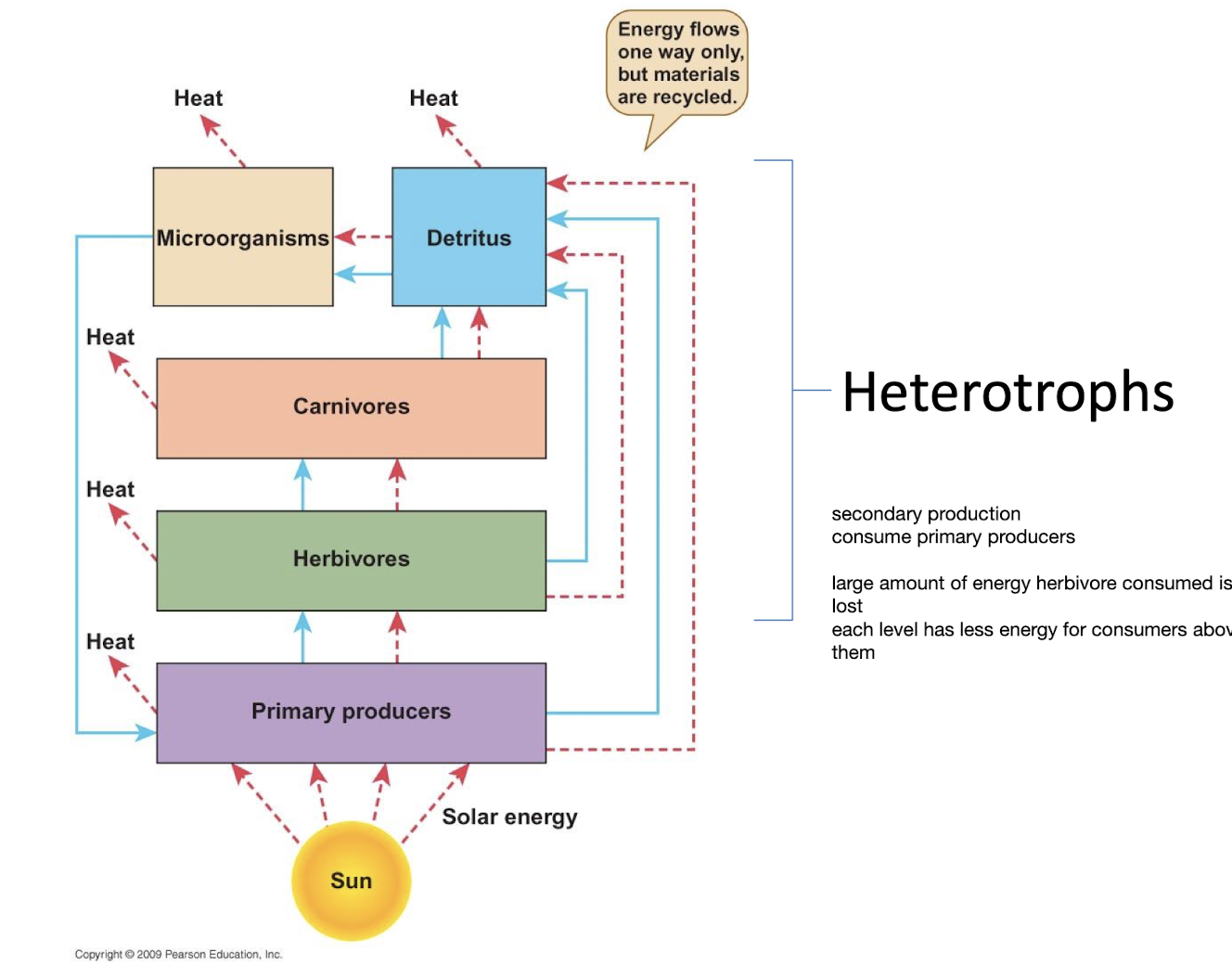
Heterotrophs
Obtain carbon from organic molecules, feed on other things like carbs proteins fats from food
secondary production, consume primary producers
lineage
chain of ancestor-descendent connections over time
Natural Selection
- Definition: The process that results from the differential survival or reproductive success of a lineage
causes heritable phenotypes with genetic basis that change in response to the environment
more effective in bringing about change in large populations
rate of this happening is an estimate of selection
consequence of this process is an adapatation
variable over time and can result in rapid evolutionary change
Fitness Wi
- Definition: A measure of reproductive success
- Types:
- Relative fitness: Absolute fitness scaled by a standard (compared to others)
- Absolute fitness: Rate of replication
adaptation
the fit between an organism and its environment
formally the heritable phenotype that increases fitness and has arisen due to natural selection in the current environment
a consequence of natural selection
can occur because there is heritable variation
xxx to one environment comes at the cost of xxx to another environment
evidence for evolution - design versus descent
all life uses the same basic inefficient materials and processes
this is only explained by the fact that we all descend from a common ancestor
common descent: children resemble their parents, we are similar and we share ancestral genes
designing an organism would make use of a wider array of processes
homologous trait
similar because of inheritance from a common ancestor. ex: human arm, seal flipper, bat wing
evidence for evolution - extinction
majority of species are extinct, species living now only represent ~1% of total biodiversity
fossils are abundant and diverse, more living species have gone extinct than are currently living
species lifespan is about 5my
ex. fossil horses first originated in asia but there were species everywhere that evolved over 55my, each species lived ~3my
evidence for evolution - adaptation
function and ancestry contribute to the evolution of the form
ex. large marine animals like whales, fish and sharks share a fusiform body shape that minimizes drag in the ocean
sharks share a CA w fish but whales are mammals descended from a hippo-like terrestrial ancestor
thus these animals share body plan through adapting to aquatic life NOT shared ancestry
Vestigial Organ
An organ or structure that has lost its original function through evolution
Adaptive Radiation
- Example: Galapagos finches from a common mainland ancestry within the last 3 my another example of their adaptations is beak length increasing after a long drought
environment acts as a potent source of selection
- Definition: The process by which organisms/a lineage diversify rapidly into several new ecological niche specialists
resource driven selection
Theory of Evolution
- Composed of many hypotheses, modification with descent resulting in a change in the genetic composition of a population, contingent and only makes use of materials available to it
- Not progressive (no species is inherently better than others)
- Involves not just selection, but also drift, migration, and mutation
- Undirected, with no intelligent designer
DOES NOT occur for good of the species
Topology
Definition: Alternative evolutionary histories represented in phylogenetic trees
Phylogeny
- Definition: A tree-like diagram showing evolutionary relationships, history of a lineage(s) (populations, genes or species)
- Key features:
• Parents/species/populations are at nodes: represent common ancestors for all descendent species
• Tips represent species
• Branches connect species, converging at nodes, trees do not depict a hierarchy or continuum
pedigree
individuals
in a sexual species, a node represents a recombined genome from two parents
any number of offspring can result
clade
a common ancestor and all its descendents
Taxon
- Definition: A named group of species, genus, order, or class
- Note: Different traits define the categories
monophyletic groups
clade, represents all species descended from a common ancestor
paraphyletic group
a group that leaves out some taxa, sharing a common ancestor
polyphyletic group
a group that includes taxa descended from multiple common ancestors
Linnean classifications
he organized species in names based on shared characters
modern taxonomy aims to construct more accurate phylogenies in terms of evolutionary history
his was NOT an evolutionary taxonomy
characters (character states)
identifiable, heritable traits
states would refer to present or absent
ancestral or derived (ex. vestigial organs appendix) NOT primitive and advanced
outgroup
species that is outside of the clade whose relationships we wish to resolve
shares a common ancestor with the monophyletic clade of interest
character states resemble those of the common ancestor
allows for a direction to be set from ancestral (shared w the xxx) to derived (not shared) on an evolutionary change
synapomorphy
shared derived character state
homologous characters because they’re inherited from a common ancestor
including gene sequences
has evolved independently in multiple taxa from a shared ancestor
is only character type that can be used to resolve phylogenetic relationships because it 1) differs from the outgroup and 2) is shared by some but not ALL taxa in the clade being studied
homoplasy
character state similarity NOT due to common descent rather caused by
convergent evolution or evolutionary reversals, or parallel evolution
so its the same character but NOT bc of evolutionary histories
Convergent Evolution
Distantly related species evolve similar traits due to similar selective pressures.
they resemble eachother more than their ancestors did
- Examples: Bats and birds, porcupines and hedgehogs (wings and spikes)
evolutionary reversals
reversion back to an ancestral character state eg. swimming in whales
especially common in DNA sequences because each site takes ¼ character states and mutation can access all of them
Parsimony
- Definition: The principle of choosing the simplest explanation or hypothesis, fewest evolutionary steps is preferred
- Application: In phylogenetics, favoring trees with fewer character state transitions
Polytomy
- Definition: An unresolved branching in a phylogenetic tree
- Usage: When we can't determine exact relationships between species
Homology
- Definition: Similarity due to shared evolutionary history
- Example: coelacanth Fish fins are xxx to tetrapod limbs as theyre more lobed and stronger
another example: mammalian ear bones are xxx to bones of reptile jaws, they evolved over time to work in the inner ear of the opossum
exaptation
natural selection co-opting a trait for a new function: ex panda’s thumb
exons
protein coding genes
under strong purifying selection because function will be preserved and tends to evolve slowly
introns
neutral in the eyes of selection and evolve faster
Parallel evolution
Similar traits evolve independently in related lineages
Bootstrapping
- Definition: A resampling technique used in phylogenetic analysis
- Process:
- Resamples the dataset multiple times
- Constructs trees from each resampled dataset
- Provides support for phylogenetic inferences
what do distance matrix methods tell us ?
lineages that are more genetically similar are more likely to be closer related
Maximum Likelihood Method
- Definition: A statistical approach for phylogenetic tree construction
- Key features:
- Uses a molecular evolution model
- Calculates probability of observing the data given a tree
- Selects the tree with the highest probability (best xxx)
bayesian methods
start w a model and a tree, change the tree slightly many times
generates probability distribution of possible trees, those with the highest probability are preferred
is HIV monophyletic?
NO! HIV evolved using convergent evolution, not a single spillover event. M,N independently evolved in chimps O,P independently evolved in gorillas, both directly from SIV. (attacks white blood cells, causing immune deficiencies)
M,N,O,P share a common mutation NOT found in SIV.
Phylogenetic Independent Contrasts
- Definition: A method for comparing traits across species while accounting for phylogenetic relationships
looks at correlation among traits based on evolutionary independent transitions
- Application:
- Used to test correlations between traits
- Helps avoid false conclusions due to shared ancestry
take the mean of two traits branching off the same node
Synonymous Substitutions (dS)
- Definition: Mutations in DNA that don't change the amino acid sequence
- Characteristics:
- Governed by chance alone
- Used as a molecular clock for estimating divergence times
often but not always selectively neutral
non-synonymous mutations
change the AA sequence of a protein and more likely to be subject to selection
neutral theory of molecular evolution
Kimura (1968) formalized it
most evolution at molecular level is NOT selective and is governed by processes associated with genetic drift
neutral mutations become fixed in lineages at regular rates
support for the theory arose from substitutions in cytochrome c gene which showed more distantly related species had more substitutions and a linear relationship
selectionists
argued the abundant genetic variation in natural populations resulted from selection
so if theres a bunch of diff niches in environment, selection will preserve the fittest type in each niche and maintain diversity
preserves specialist in each niche
sometimes called balancing selection
neutralists
believe that most genetic variation was selectively neutral, did not impact fitness
Selective Sweep
Definition: The process by which a beneficial mutation rapidly increases in frequency in a population, a beneficial allele fixes faster than a neutral allele
- Example:
- D614G mutation in SARS-CoV-2 spike protein
genetic hitchhiking
mutations that are linked physically to the selected allele ‘hitchhike’ to high frequency
Purifying Selection
- Definition: A type of natural selection that removes harmful mutations
- Indicator:
- dN/dS ratio less than 1
non-synonymous substitutions (dN)
change protein, faster evolution
Tajima’s D
uses two estimates of genetic diversity to ask whether a population is evolving neutrally or not
S = number of variable sites
pi = mean number of differenences between a pair of sequences
these both estimate theta = the quantity of genetic diversity in a population under neutrality
D = thetapi - thetaS
tells us whether a population is evolving neutrally or not
D = 0 is neutral
D < 0 = directional selection( more rare alleles than expected, suggests selective sweep eliminated variation happened recently)
D > 0 = balancing selection (fewer rare alleles than expected, suggests selection favours distinct alleles in the same population)
Macroevolution:
• Starts with speciation
• Involves large-scale evolutionary changes over time
considers broader changes in diversity at higher taxonomic levels and how this is distributed
microevolution
changes in allele and gene frequencies within populations
speciation
how genetic changes within populations lead to new evolutionary clumps
defined in terms of reproductive isolation = will thus create a new species
biogeography
study of the distribution of species across space and time
dispersal
movement of populations from one region to another with limited or no return exchange ex. marsupial evolution : most living are found in Australia but oldest fossils were found in china and north america evolved w a mix of this and the other because the modern day marsupial has CA from N america and S america too
vicariance
formation of geographic barriers to dispersal that divide a once-continuous population. ex. if continent divides and population splits there could be more species so for marsupial their phylogenetic patterns mirror the order in which the continents broke up
anagenesis
wholesale transformation of a lineage from one form to another
not our understood definition of evolution, things did not evolve gradually, there were species that took over eachother
punctuated equilibria
periods of stasis followed by brief periods of rapid morphological change linked to speciation
sees speciation and morphological change happening simultaneously
ex. bryozoans diversification pattern works well with this model
gradualism
slow, gradual morphological changes over time resulting in speciation
involves anagenesis and speciation
turnover
number of species eliminated and replaced per unit time
standing diversity
number of species present in an area at a given time
ecological opportunity
presence of vacant niche space
absence of competitors opens this up for a lineage
leads to adaptive radiation
key innovation
trait(s) that allows a lineage access to new resources
evolutionary novelty
new genetically based trait
used to improve in totally new conditions, take whatever enzymes you have and grow faster
EAD model for evolution of novel gene function
Exaptation: use what you have to perform a new role
Amplification: make more of what you have (enzymes) even if its not the best
Divergence: improve fitness through changes in gene function, more copies of genes allows one copy to diverge without compromising og function
promiscuous proteins
exaptations with one main function and many side functions as well
sloppy which makes organism get by in an environment where its not well adapted
paralogs
genes descended from duplications
evolution of novel trait in e coli example
usually cannot metabolize citrate aerobically (diagnostic trait)
a citrate metabolising strain emerged!
exaptation: e. coli had the machinery to do this but it previously only worked anaerobically
amplification: duplication of citT and linked sites inserted after the promoter allows for its expression in oxygen presence
divergence: intitially strain could barely grow but further copies accumulated mutations that led to citrate specialist that coexisted with glucose specialist
cambrian explosion
sudden appearance of diverse animal body forms ~541 mya
thought to have originated from early developmental mutations leading to new body plans
caused by changing geology expanding habitats
new ecological opportunity
background extinction
the normal rate of extinction for taxa or biota
mass extinction
a statistically significant increase above background extinction rates
origination
when the rate of new species formation exceeds the rate of extinction
leads to an increase in diversity
cladogenesis
the splitting of a lineage into 2 or more descendant lineages
mutation
any change to the genetic sequence, will be inherited by the cell’s descendants, imperfect copying procedures
Only way to introduce new variation into a population
- Somatic ones are not passed on to offspring
- Insertions and deletions (indels) can affect gene function
rates for any given gene are low but per genome and population many new ones arise each generation, which will generate variation
Chromosomes
Condensed DNA strands wrapped around histones
Plasmids
Can move independently of host, potentially spreading antibiotic resistance
microRNA (miRNA)
can block translation and have major phenotypic effects, non-coding RNA that regulates gene expression
alternative splicing
RNA splicing to create multiple proteins from a single gene in eukaryotes
ploidy
number of copies of unique chromosomes in a cell
Gene Duplication
- Can change physical appearances significantly
- Maintains ancestral function while diverging to create novel functions
- More genetic material provides more mutation material for natural selection
thought to explain lots of rapid diversification and rise of new clades
the c-value paradox
the amount of DNA in a haploid genome does not correspond to the complexity of the organism
rather its mostly pseudogenes and mobile genetic elements that parasitize the host’s replication machinery to make copies of themselves
somatic mutations
affect cells in the body of an organism; not heritable but they can be very damaging
germ-line mutations
affect gametes; heritable and relevant to evolution, other type isnt
Conditional Expression
Some traits (e.g., beetle horns) only appear after reaching a certain body size or nutritional threshold
Heritability
How much of a population's variation is associated with genetics
the proportion of the phenotypic variance that is attributable to genetic differences among individuals
a ratio of variances
fraction of phenotypic variation that is passed to the next generation
what does Independent Assortment (IA) do?
Determines which chromosomes go into which gamete
Recombination
Creates variation different from parents but not brand new
genotype
the genetic makeup of an individual
phenotype
an observable, measurable characteristic as the manifestation of the genotype of an organism (PHYSICAL)
P = genotype + environment
ex. human height depends on resources and nutrients along with internal morphogen signalling molecule gene expression exchange rate
polyphenic trait
single genotype produces multiple phenotypes depending on environment
quantitative trait locus QTL
examines known markers across the genome to identify those that are linked to high/low values of a trait
markers closely linked to genes conferring a trait value are likely to remain linked despite recombination
marker is not usually the gene of interest
use log of odds (LOD) score to indicate an association between a marker and a trait value that is unlikely to have arisen from chance
population genetics
studying the distribution and frequencies of alleles in populations and the mechanisms driving allele frequency change
how genes move through populations
population
a group of interacting and potentially interbreeding individuals of the same species
group of organisms of the same species occupying a particular space at a particular time
usually collections of smaller sub-xxx connected through dispersal
extent of subdivision depends on features of landscape and motility of individuals
if isolated it becomes genetically distinct
drift occurs independently, different alleles will be lost and genetic divergence overtime occurs in each sub-xxx
genetic locus
location of a specific gene or sequence of DNA on a chromosome
could be allele of gene, segment of genome, or specific nucleotide etc
deliberately vague
haploid
individuals carrying a single copy of an allele at each locus
diploid
individuals carrying two alleles at every locus
homozygous
AA alleles are the same
heterozygous
Aa alleles are different, have greater fitness than the other only in diploids as haploids don’t have another chromosome
Null Model
A model where no evolutionary change is expected
- No natural selection, migration, or other drivers of evolutionary change
- Important because it sets a baseline for comparison
any deviation from this model tells us the population is evolving even tho we may not know the cause