L3: Eukaryotic chromosome replication I
1/80
Earn XP
Description and Tags
Replication origins, centromeres, telomeres
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
81 Terms
How does chromosome replication differ in eukaryotes to prokaryotes
replication initiates at many sites per chromosome
one round of rpelication and cell division is completed before the next starts
parental & progeny DNA is assembled into nulceosomes and higher order chromatin strucutres
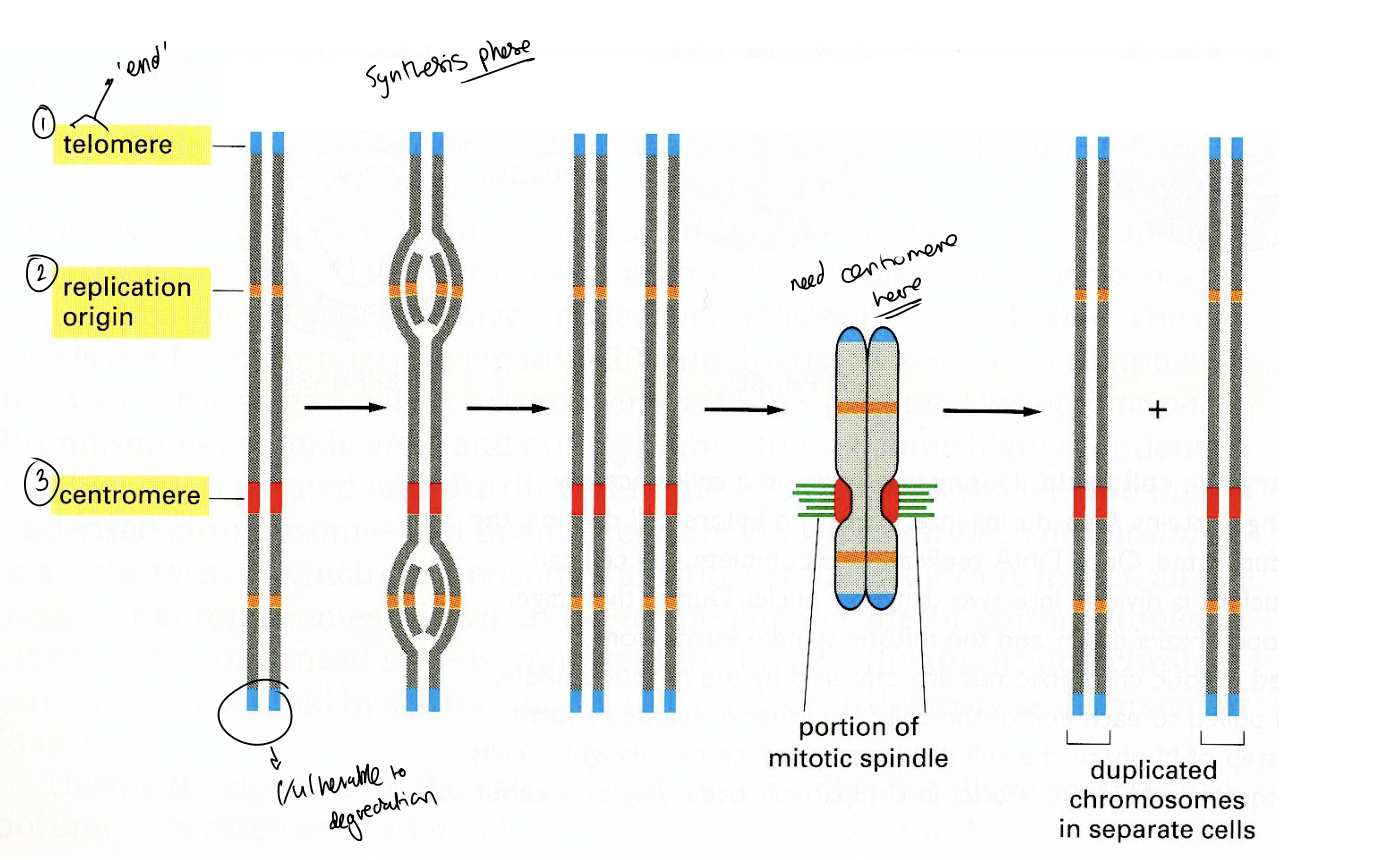
The three important elements on chromosomes for DNA cycle
Relication origin
Centromere
Telomere
What are the DNA replication initiation sites called
Replication origins
Why is it useful for origin to be specific sites
so that replication cannot just keep happen all the time
need to be specific r=time at specific sites
helps to regulate it
what we know about replication origins in eukaryotes?
We know more about prokaryotic, animal viruses and lower eukaryotes (e.g yeast0 replication origins
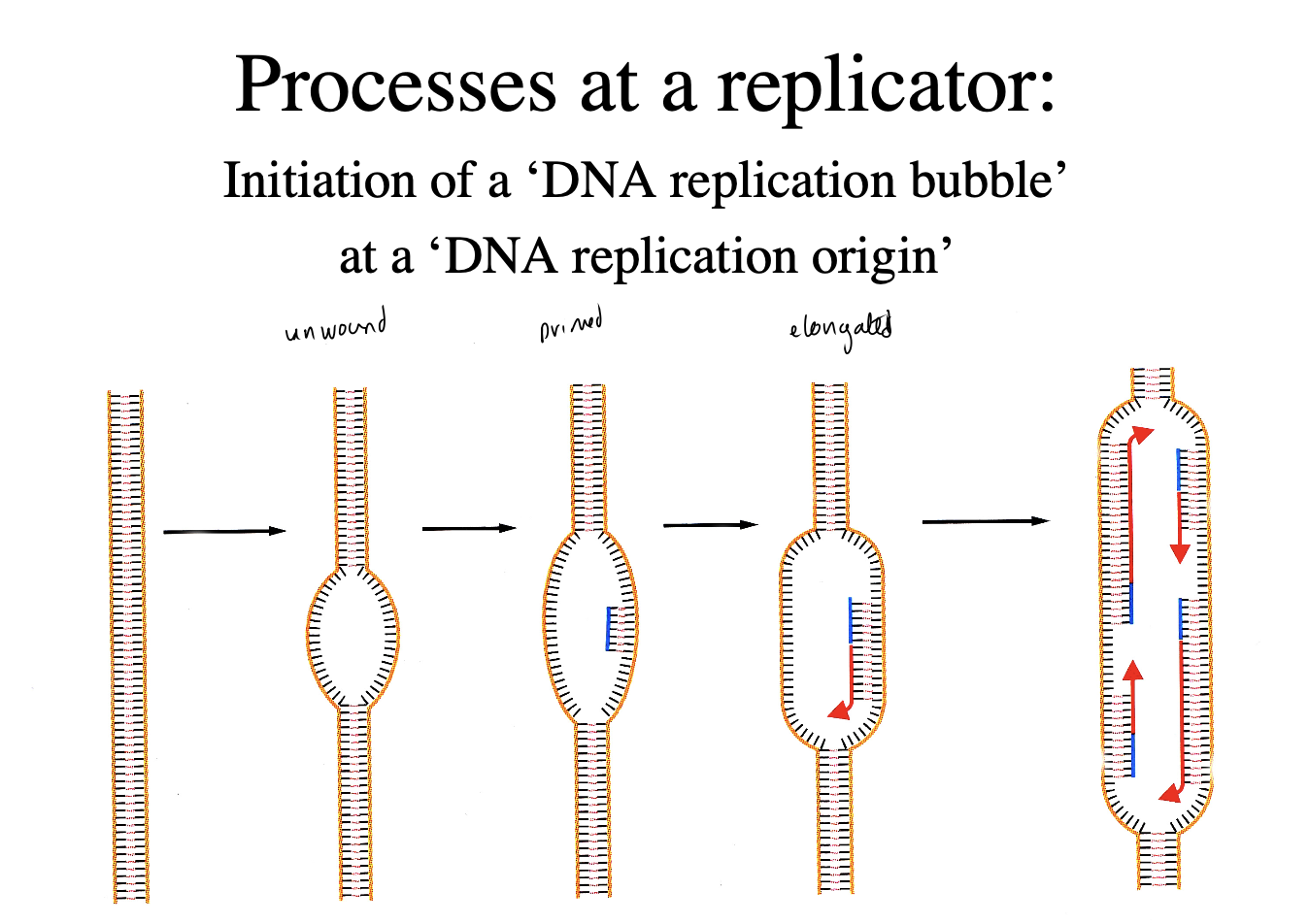
The replicon model: start with a conceptual idea of how replication of circular bacterial chromosomes might be regulated
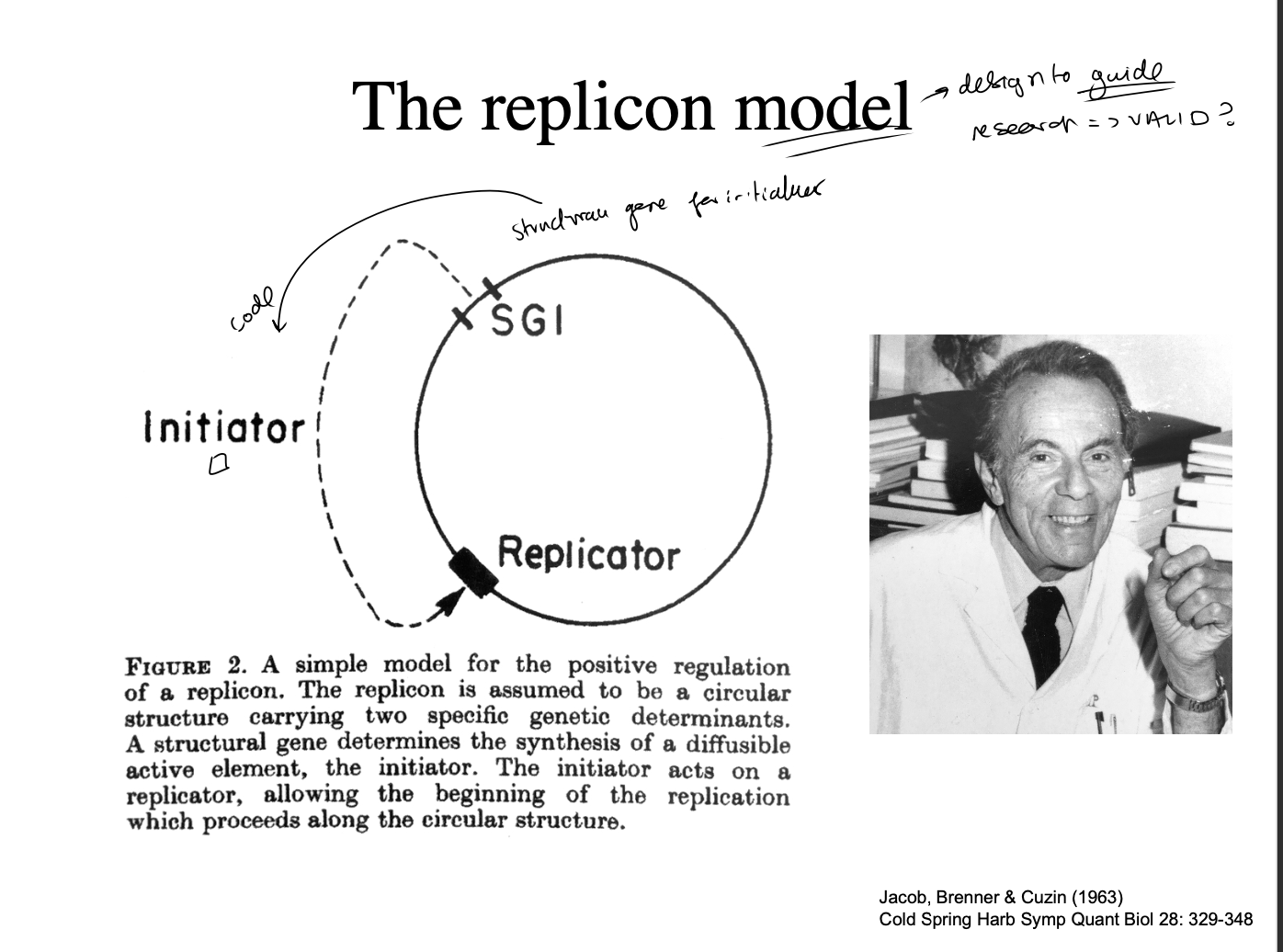
Processes at a replicator
unwound
primed
elongated
where is knowledge of higher eukaryotic origins from?
biochemical
genetic analyses of few example cases
high thorughput DNA sequencing analyses at genome-wide resolution
Is the model supported?: Replication origins of animal viruses: they do nothave all the elements
e.g SV40 and polyoma virus
organised as minichromosomes
what do they have→ only the control elements for their own replication:
origin sequences
initiator protein
Imagese of replicating circular SV40 DNA

How do they get the other factors required for replication?
recruited from the infected host cell (monkey or human)
therefore:
they becaome the first simple and useful model systems for eukaryotic chromosome replication
What is the initiator protein
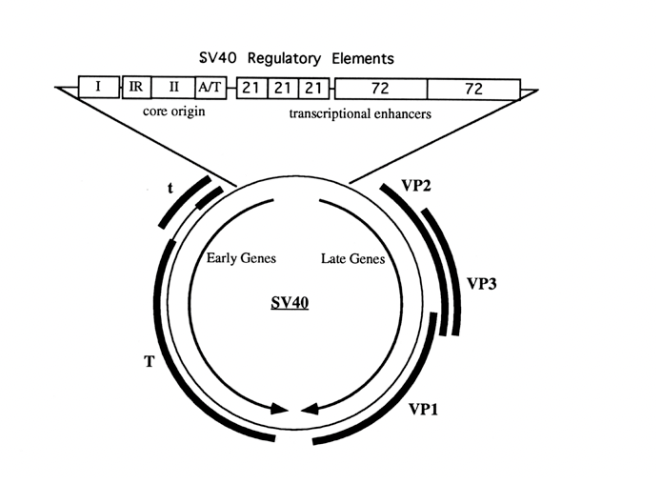
‘Large T antigen’
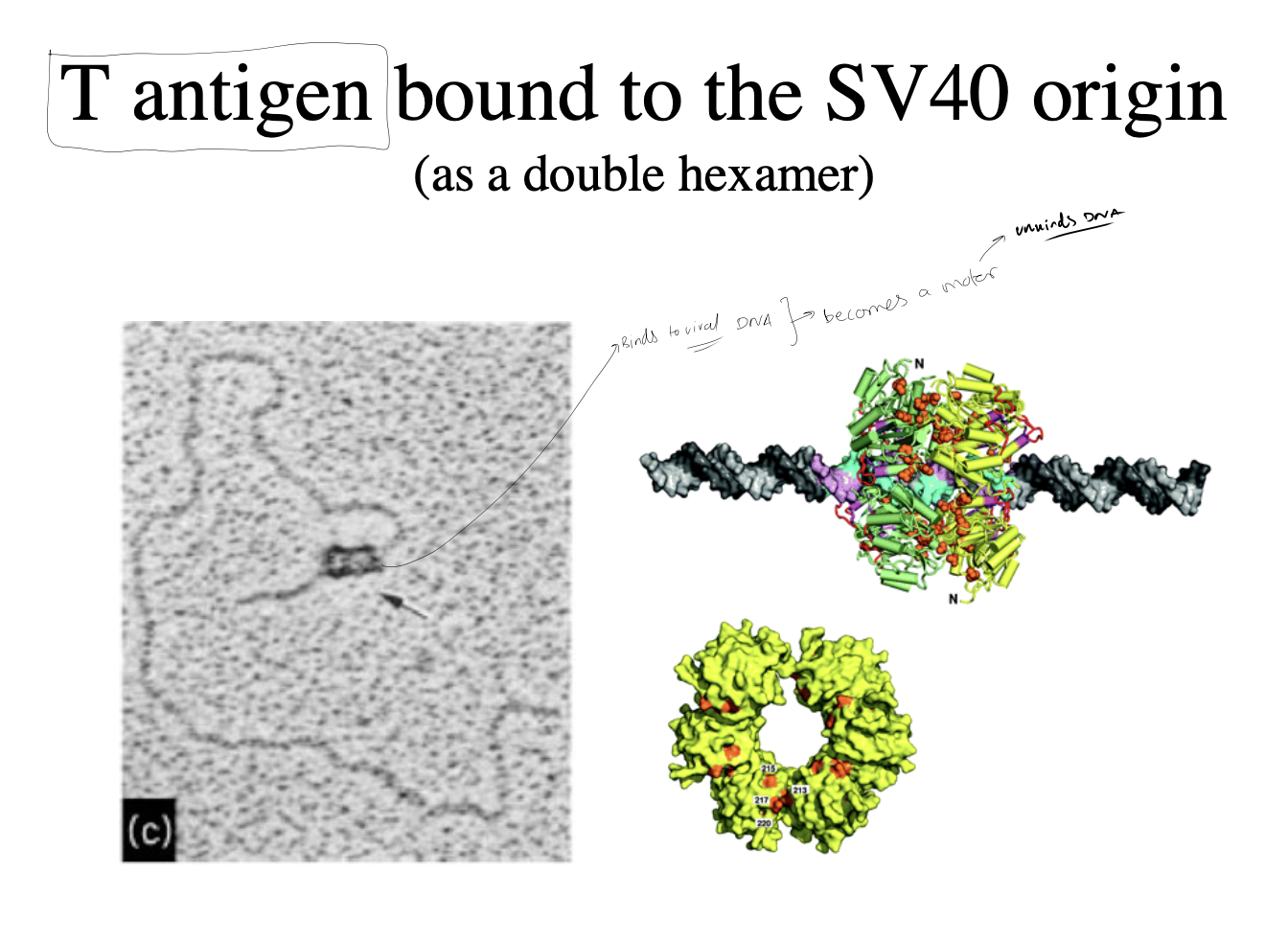
What does it do to initiate replication
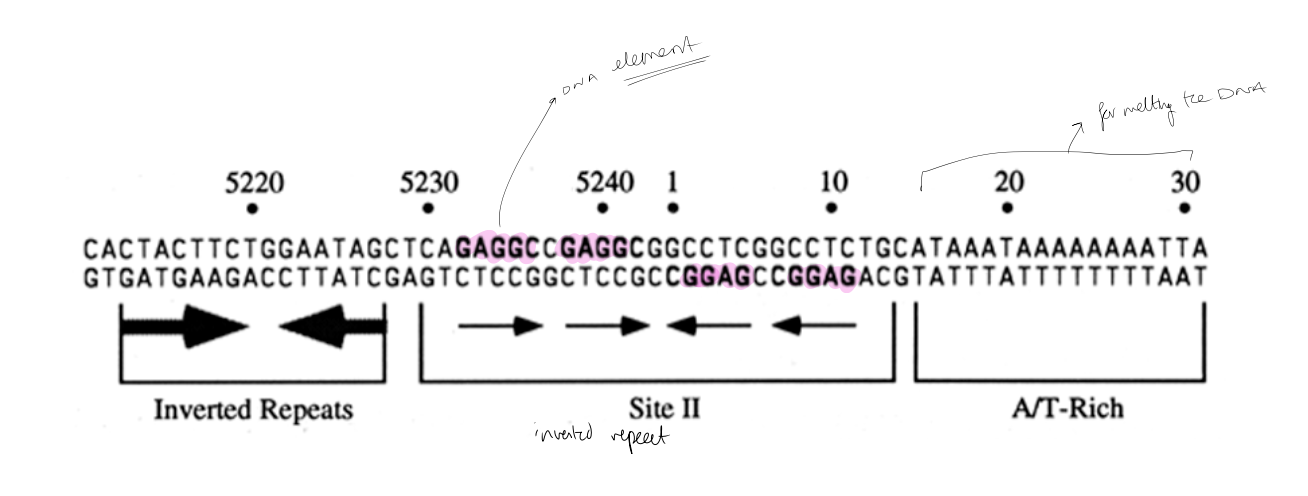
bind to unique site on viral genome
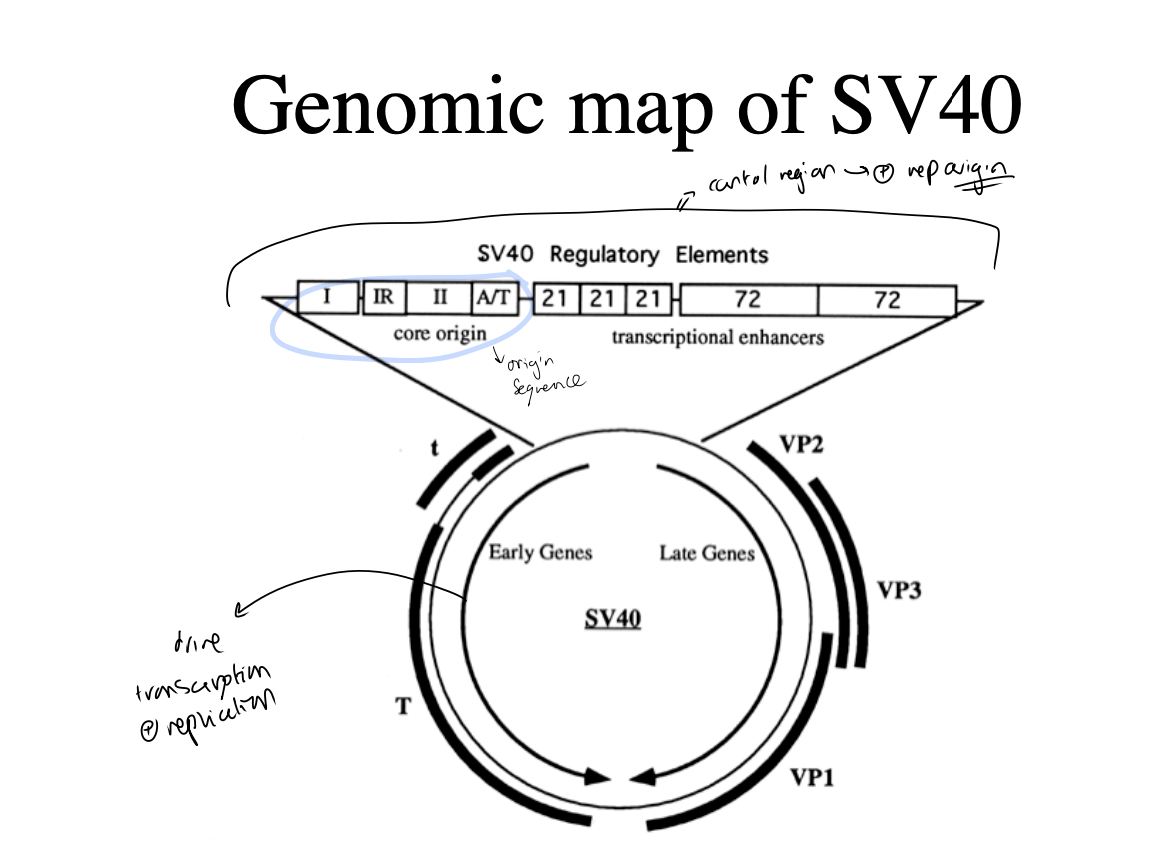
65bp control region containing:
a 27 bp inverted repeat (a potential hairpin)
conserved A/T rich element is needed for origin function
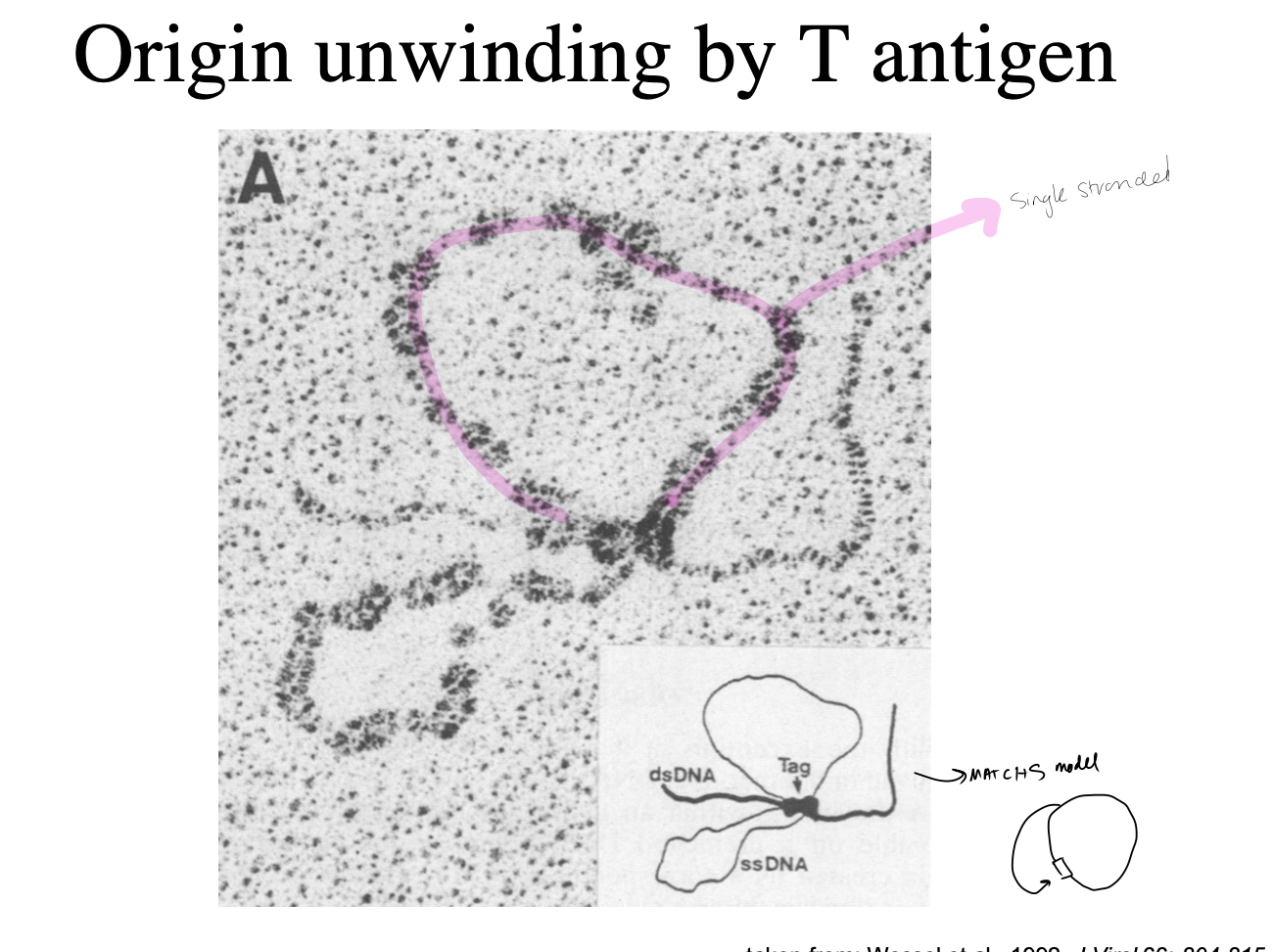
Origin unwinding by T antigen
see the single stranded DNA is tagged with proteins in the EM
→ THEREFORE: this matches the replicon model
Overall genomic map of SV40
Large T antigen (early genes)
Late genes
Control region + replication origin with origin sequence
Why are virus origins not valid models for cell chromosomal orgins?
virus must replicate many times in one cell cycle for propagation
→ therefore: defies cellular controls
Replicating chromatin image (drosphila)

How are chromosomal origins different
Linear chromosomes
Many origins per chromosome
activated only once per cell cycle
Yeast: plasmids
2 mu plasmid
organised as spisomal minichromosomes
Yeast plasmids: what does they replication require
specific DNA seqeunce element→ autonomously replicating sequencse ARS element
What does the ARS element act as
replication origin
allows initiiation
ARS elements were isolated from random genomic DNA fragments by…
their ability to confer the ability to replicate
as an autonomous plasmids in yeast cells→ after ligation to an origin-less circular plasmid
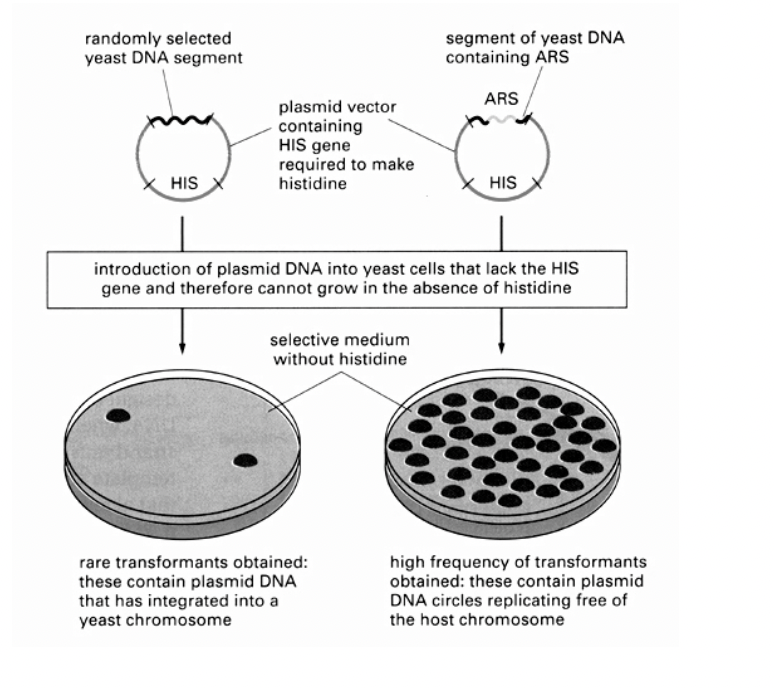
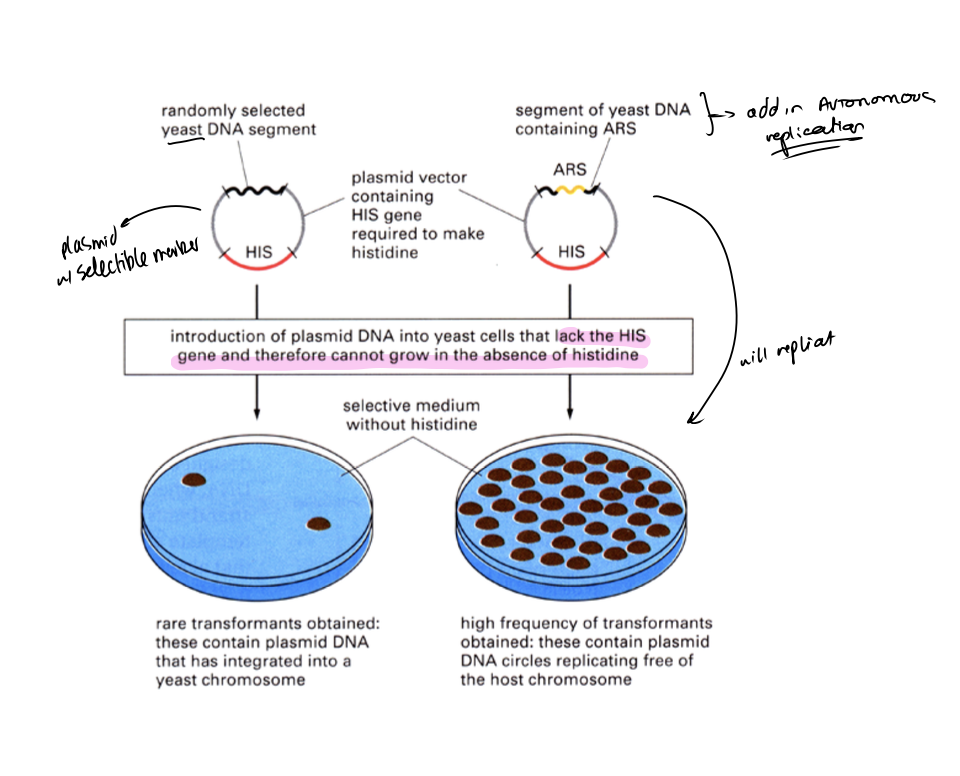
How experiment worked
Get a plasmid vector containing a selectible marker
i.e HIS gene required to make histidine
Add randomly selected yeast DNA segment
If does not contain ARS→ rare transformants obtained: these contain plasmid DNA that has integrated into a yeast chromosome
some will be able to survive but the offspring of this will not have the plasmid so will not→ that is why you get SOME colonies but not all of them
If it does contain ARS→ high frequency transformants obtained
contain plasmid DNA circles replicating free of the host chromosome
Mutant analysis: Deletion and point mutants in typical ARS element define…
an essential consensus
A box
for ARS activity
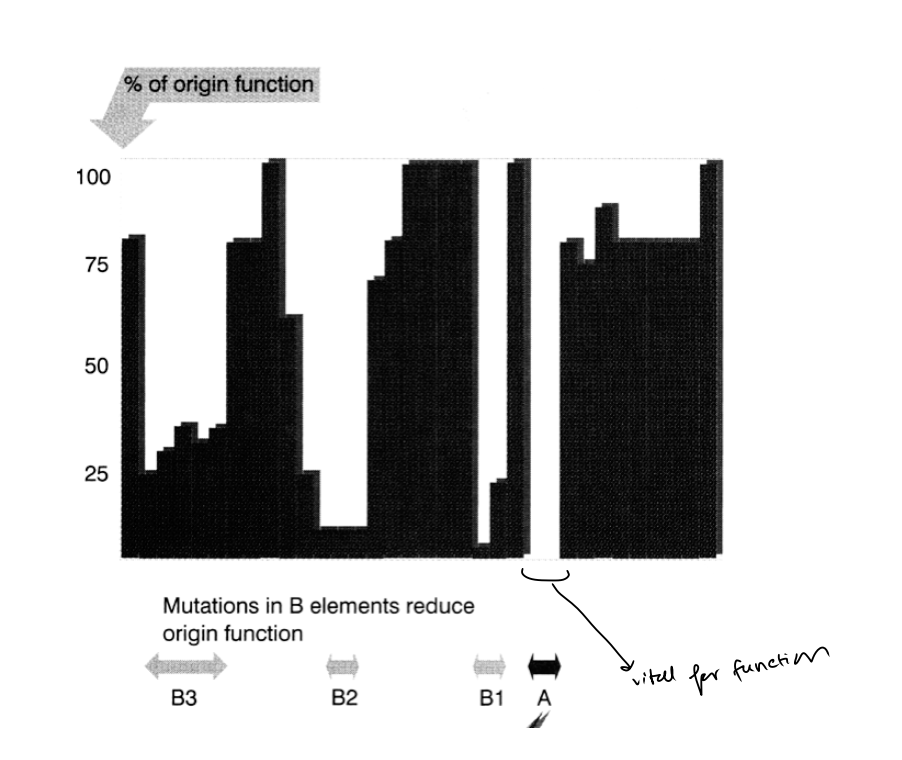
Mutant analysis: What are B elements
Flanking sequencing
affect the efficieny of ARS function
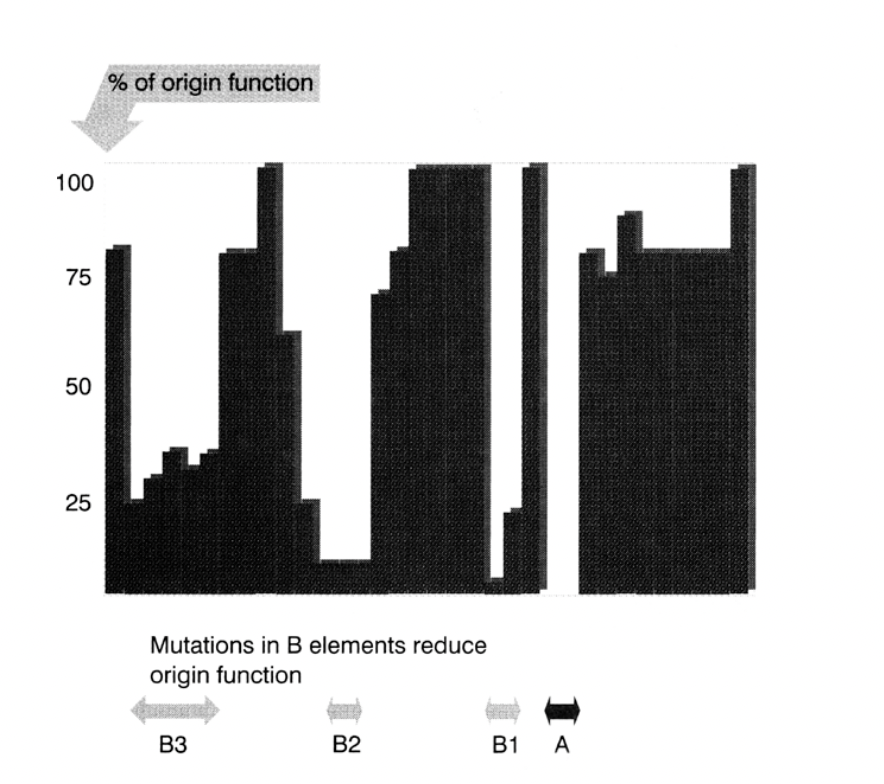
What does this mutant analysis overall show
Get an idea of wchih parts are more important than others
which parts can/cannot stand mutation
OVERALL: Gives ideas of the spacing of components along the ARS
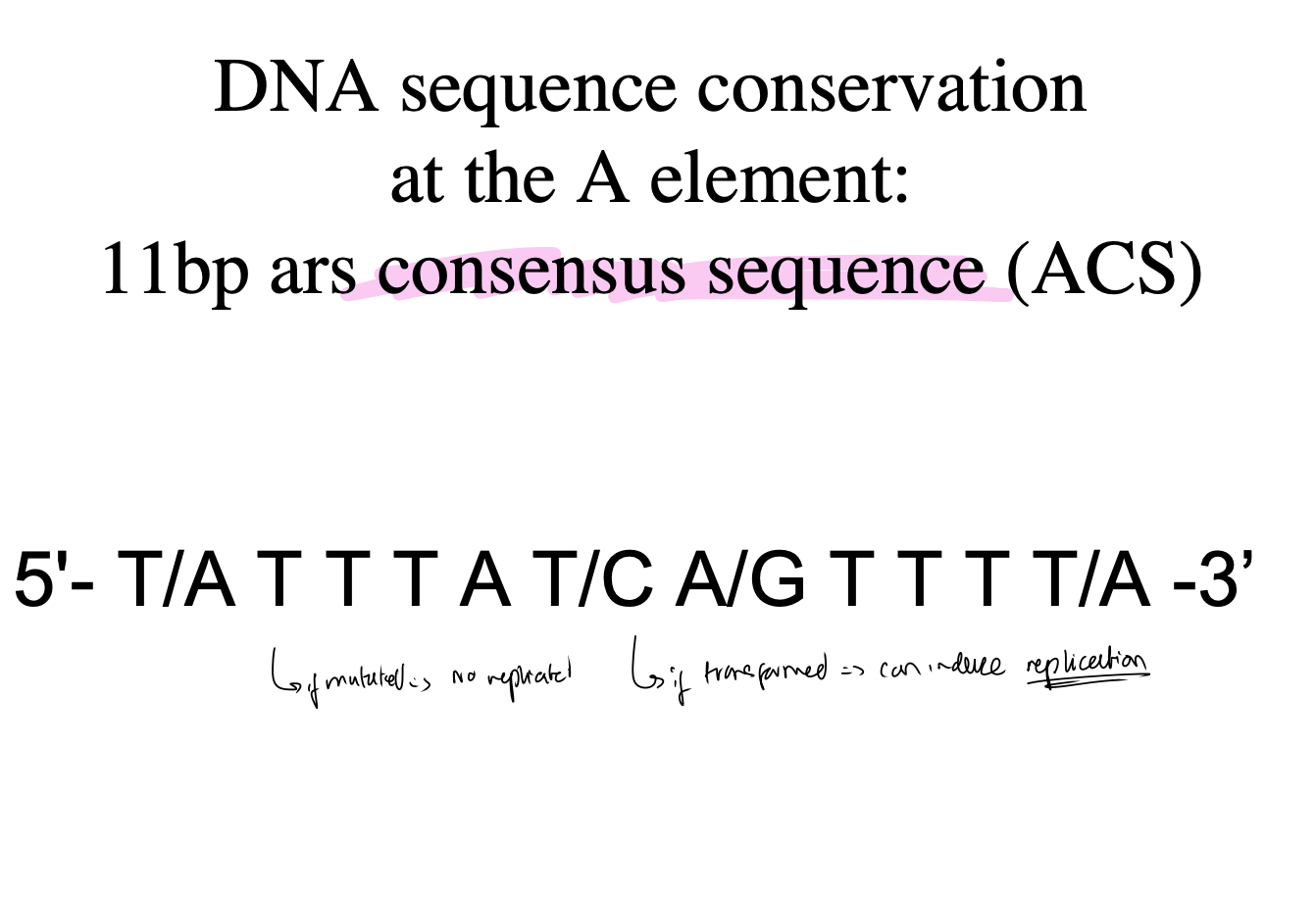
What is the DNA sequence conservation at the A element:
11bp consensus sequence (ACS)
How many ARS elements are found in chromosomal DNA of budding yeast
over 200 ARS elements
In this context, they also function as
origins
However…
replication does not always initiate at all possible yeast origins
How are the A and B elements not degraded by nnuclease?
Experiment: with nuclease digestion in vivo
show DNA sequences are protected by associated proteins
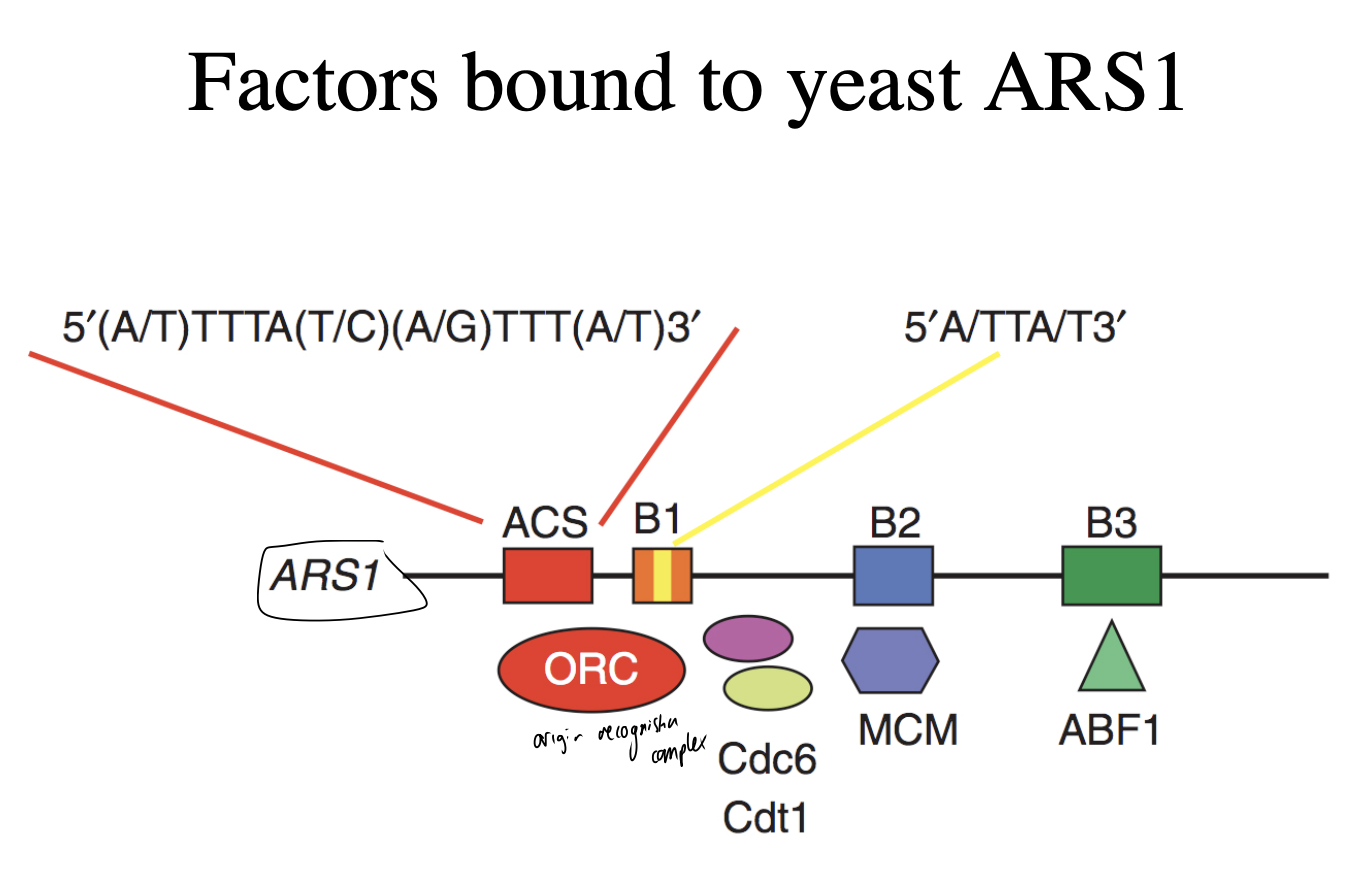
How are yeast origins recognised
sequence-dependent manner
by initiator protein complex
What is the initator complex called
origin recognition complex ORC
How does the ORC work
This diagram shows the spacing that the mutant analysis hinted at
recognises origin
binds with A box in presenece of ATP
cdc6, cdt1, MCM complex associate with ORC in a stepwise manner
occupying additional positions e.g B elements
overall: forms the pre-replicative complex
Why is yeast a good model
Small
acts like bacteria→ colonies, plasmids, etc
Eukaryotic
5-6 thousand genes
analagous to human genes
humans: 20000 genes→ will be duplicates and divergents of the yeast ones
Replication origins in higher eukaryotes: human chromosomes are big, meaning with two divergent forks
take a month to replicate from a single initiation site
moving at 3kb/min
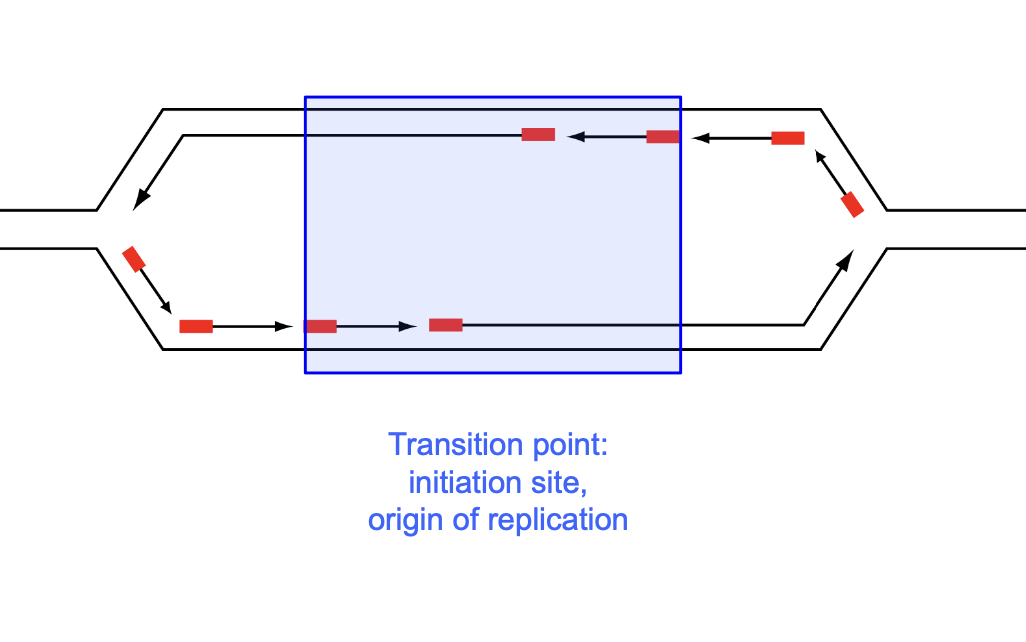
Features in mammalian replication:
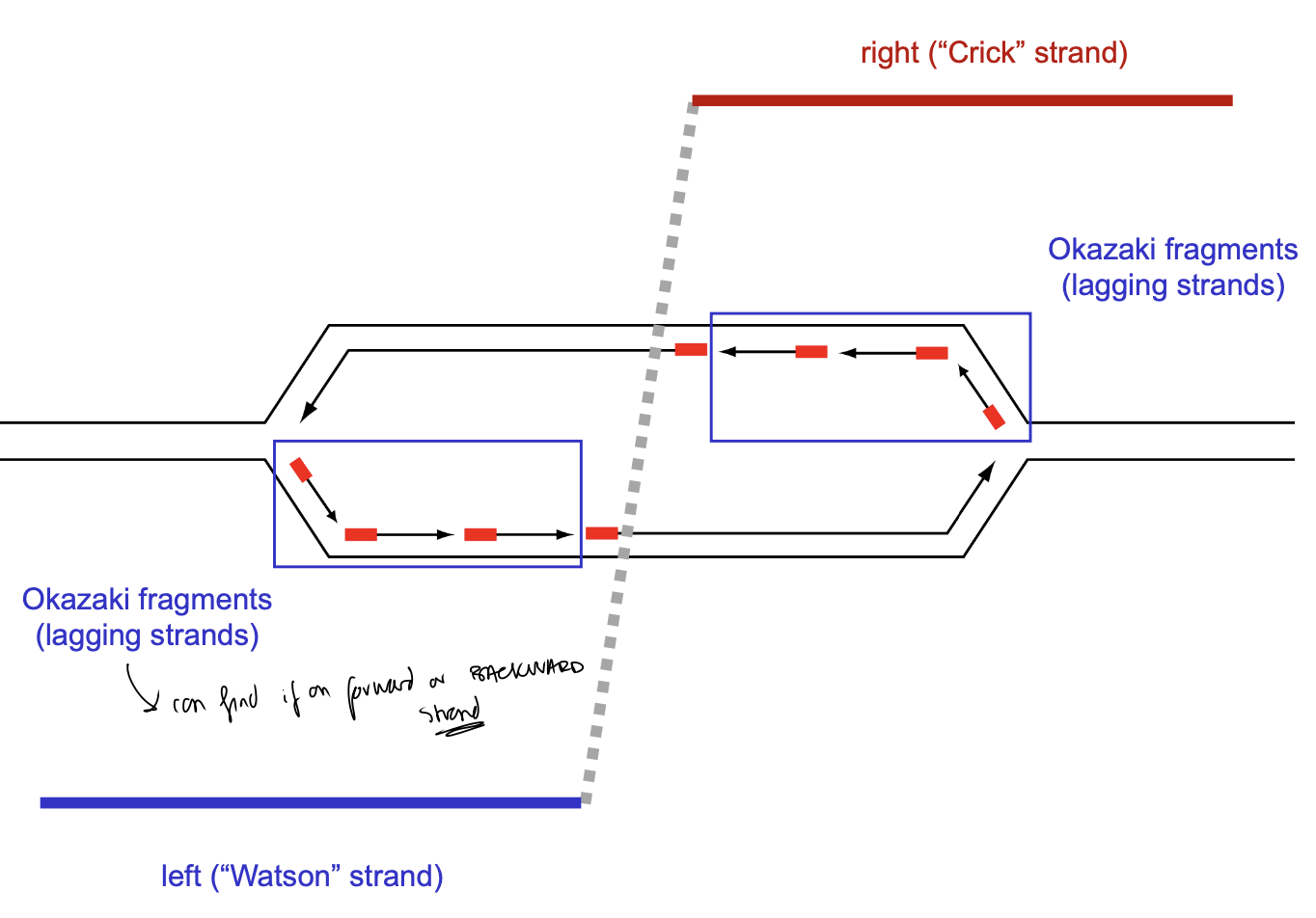
okasaki fragments
Transition point: initiation site, origin of replication
Forward nascent strands (leading strands)
How come replication only lasts less than 10 hours?
have several thousand replication origins present per chromosome
tens or hundres of thousand origins per genome
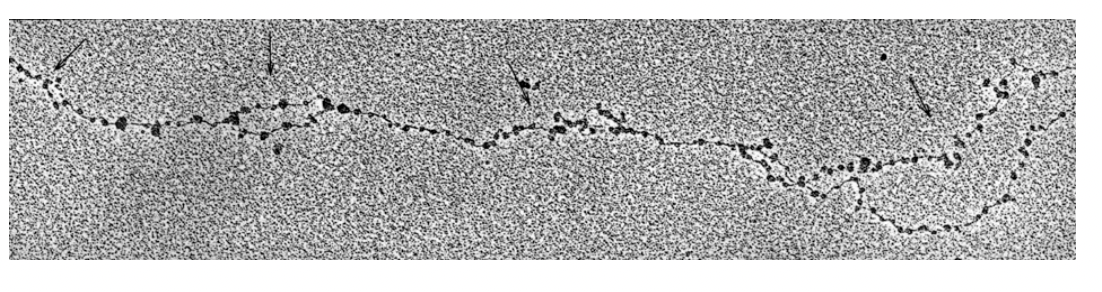
EM analysis of replicating chromosomes showed that
there are replication bubbles emanating from activated origins
at intervals of 30-300kb
Site specific replication origin is defined and mapped at high precision by
determining the transition point
between leading and lagging strand synthesis
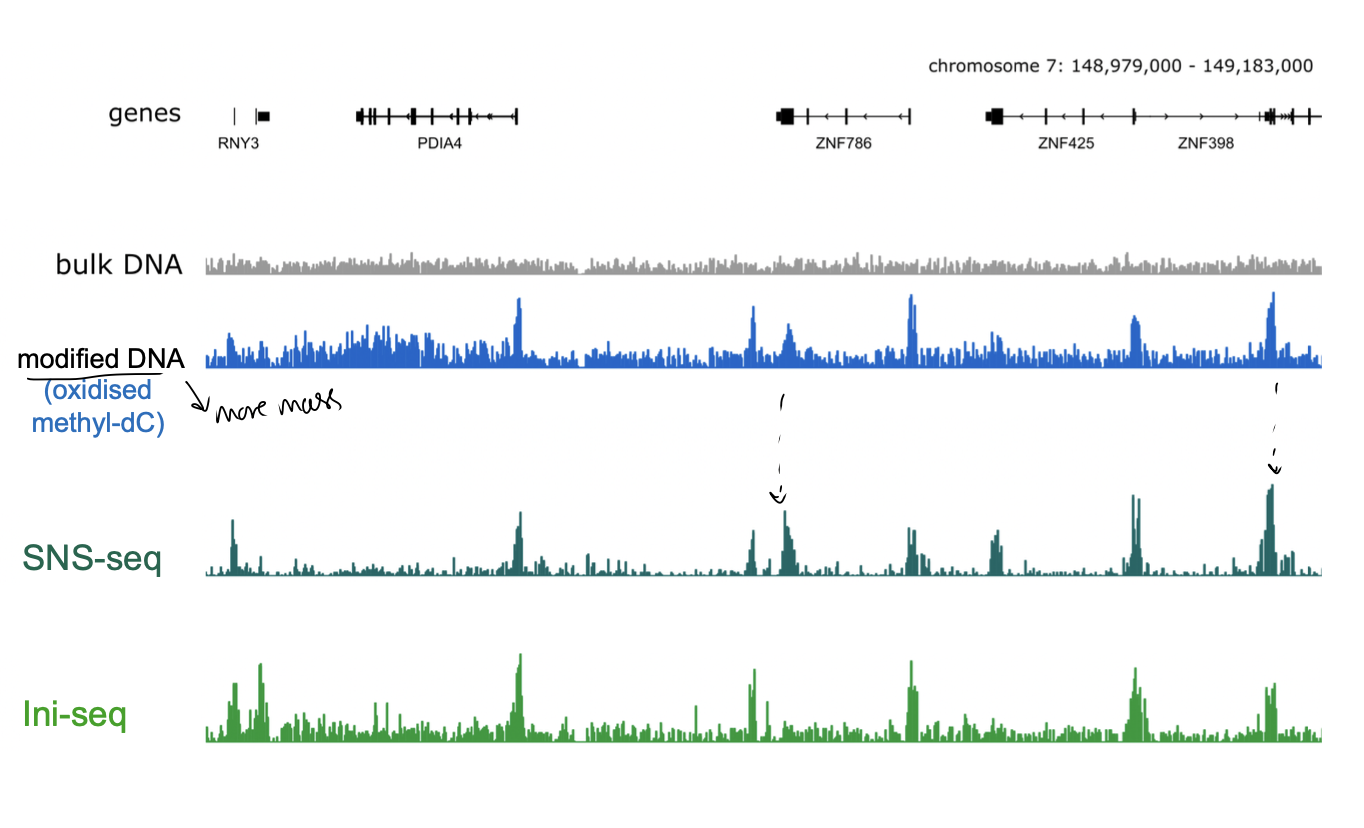
What technologies have made mapping DNA replication origins in vertebrate cells easier?
high-throughput DNA sequencing
e.g SNS-seq
e.g ini-seq
computational analyses
identified some tens or thousand potential origin sites in the human genome
Genome-wide origin mapping techniques
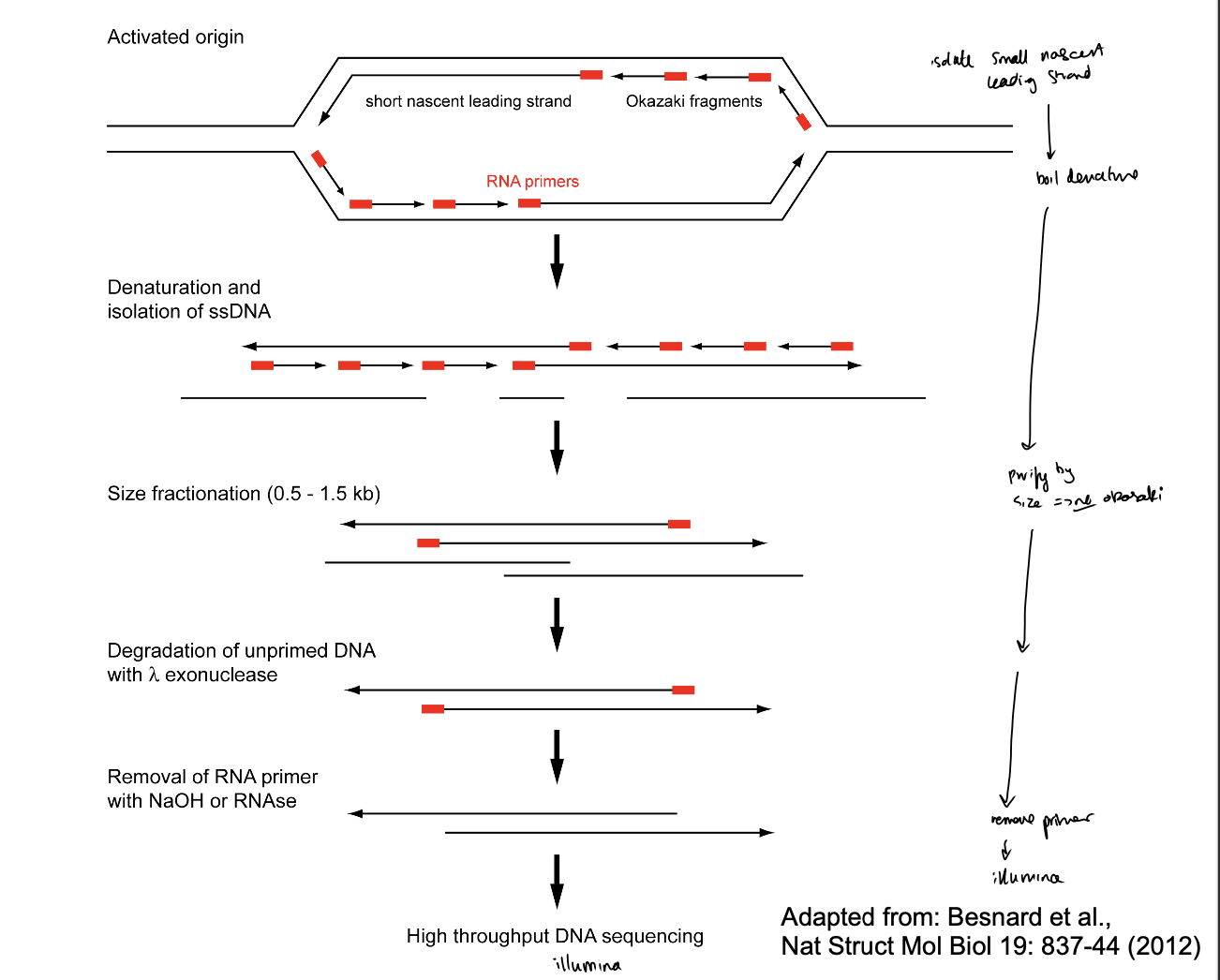
Sequencing of short nascent strands (SNS-seq)
Sequencing of initiation sites (Ini-seq)
Sequencing of Okazaki fragments (OK-seq)
What is SNS-seq
small nascent DNA leading strands tagged so know they are new strands that have been replicated
isolated from replicating cells
sequenced to localise replication origins
Also a bulk experiment
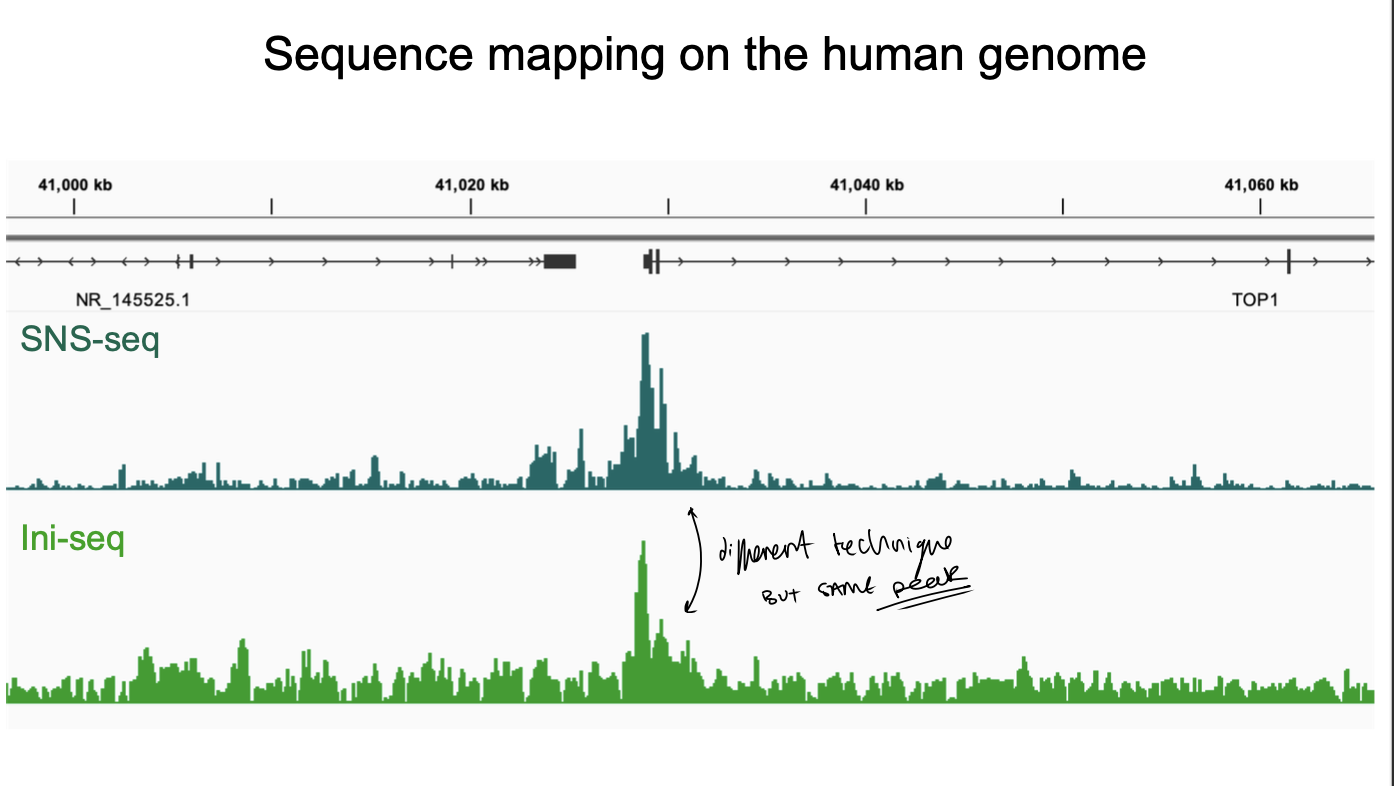
SNS-seq sequence mapping on the human genome
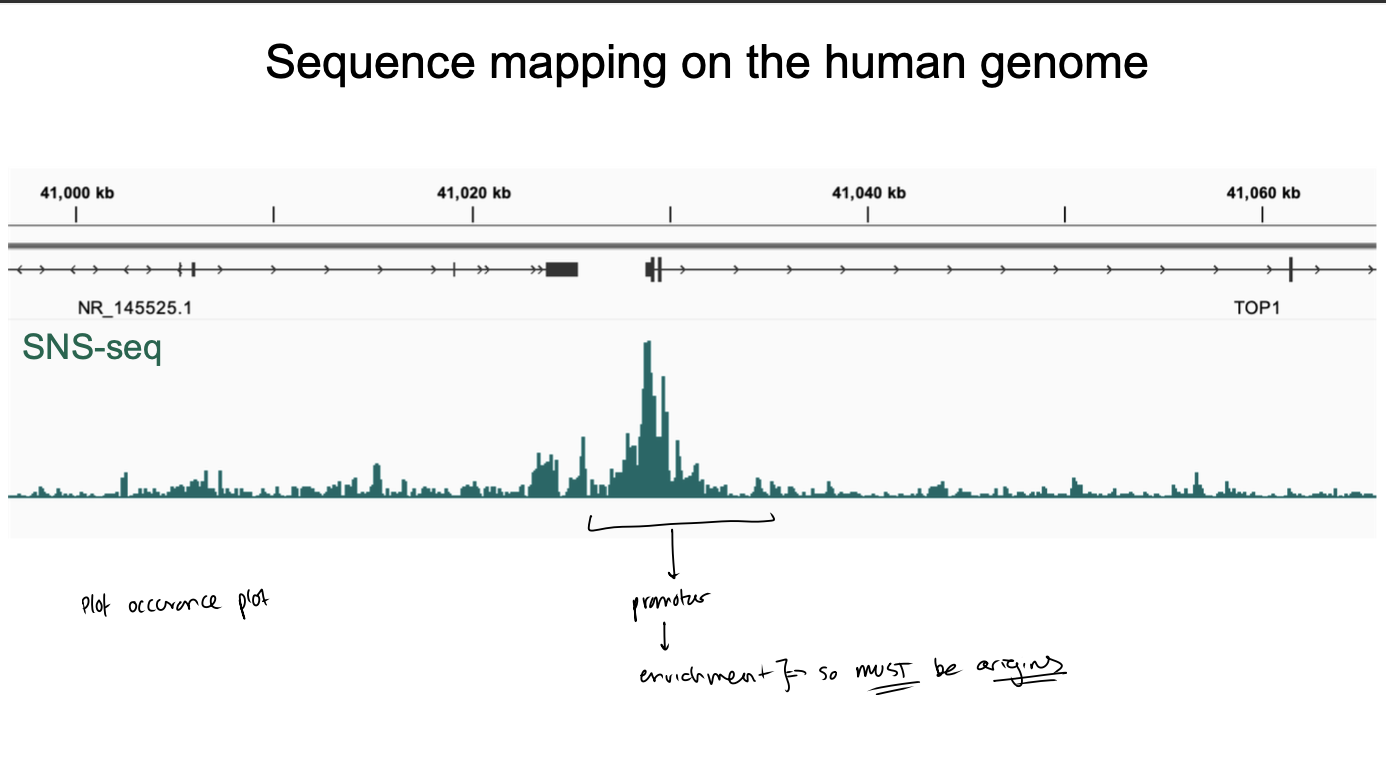
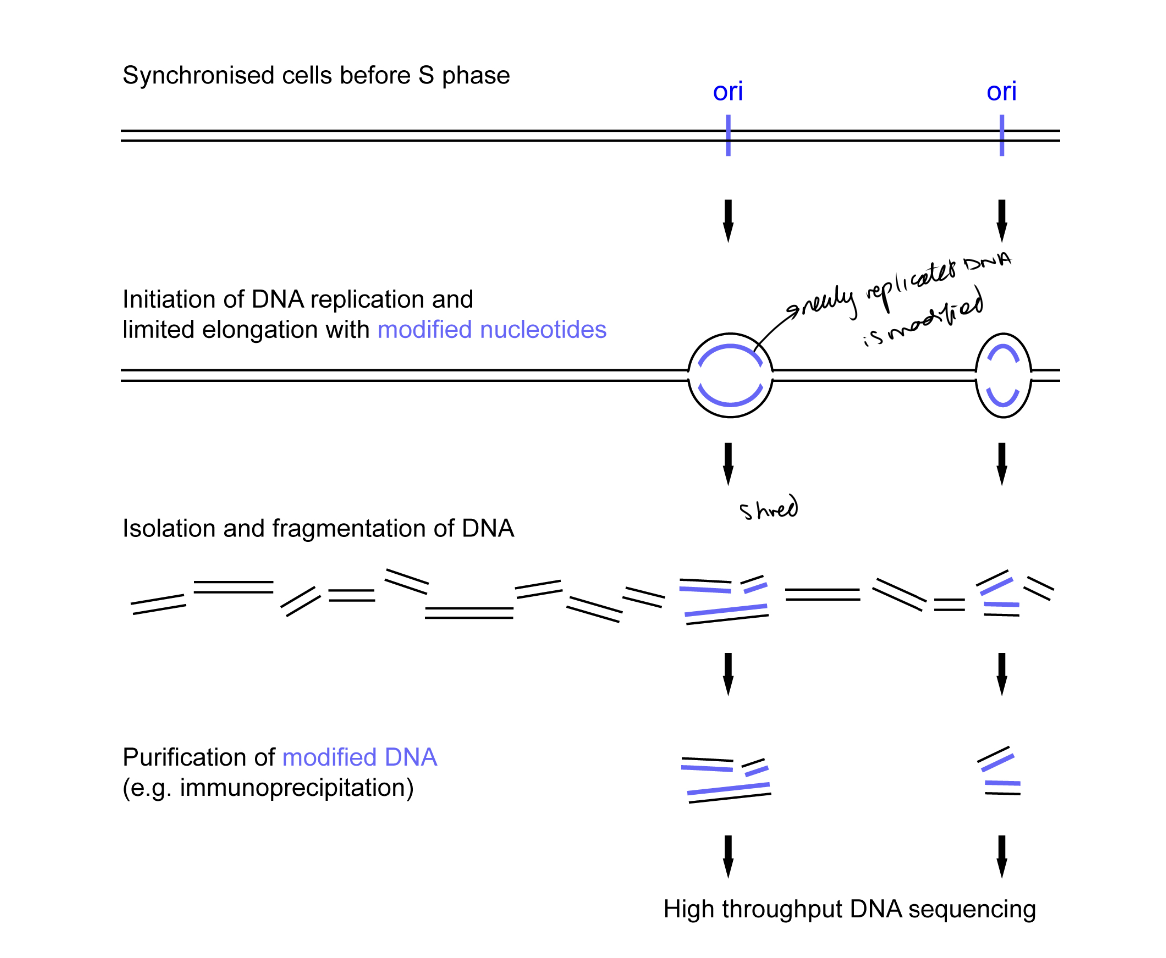
What is ini-seq
DNA replication initatied in nuclei of cells synchronised
in late G1 phase of cell cycle (just before DNA rep)
DNA allowed to replicate only for a short time following initiation
labelled by modified nucleotide
initiation-site associated replicated DNA is isolated and sequenced
SNS-seq and ini-seq have both
yielded consistent result
detailing tens of thousands of defined replication origins in the human genome
OK-seq: Okazaki fragments
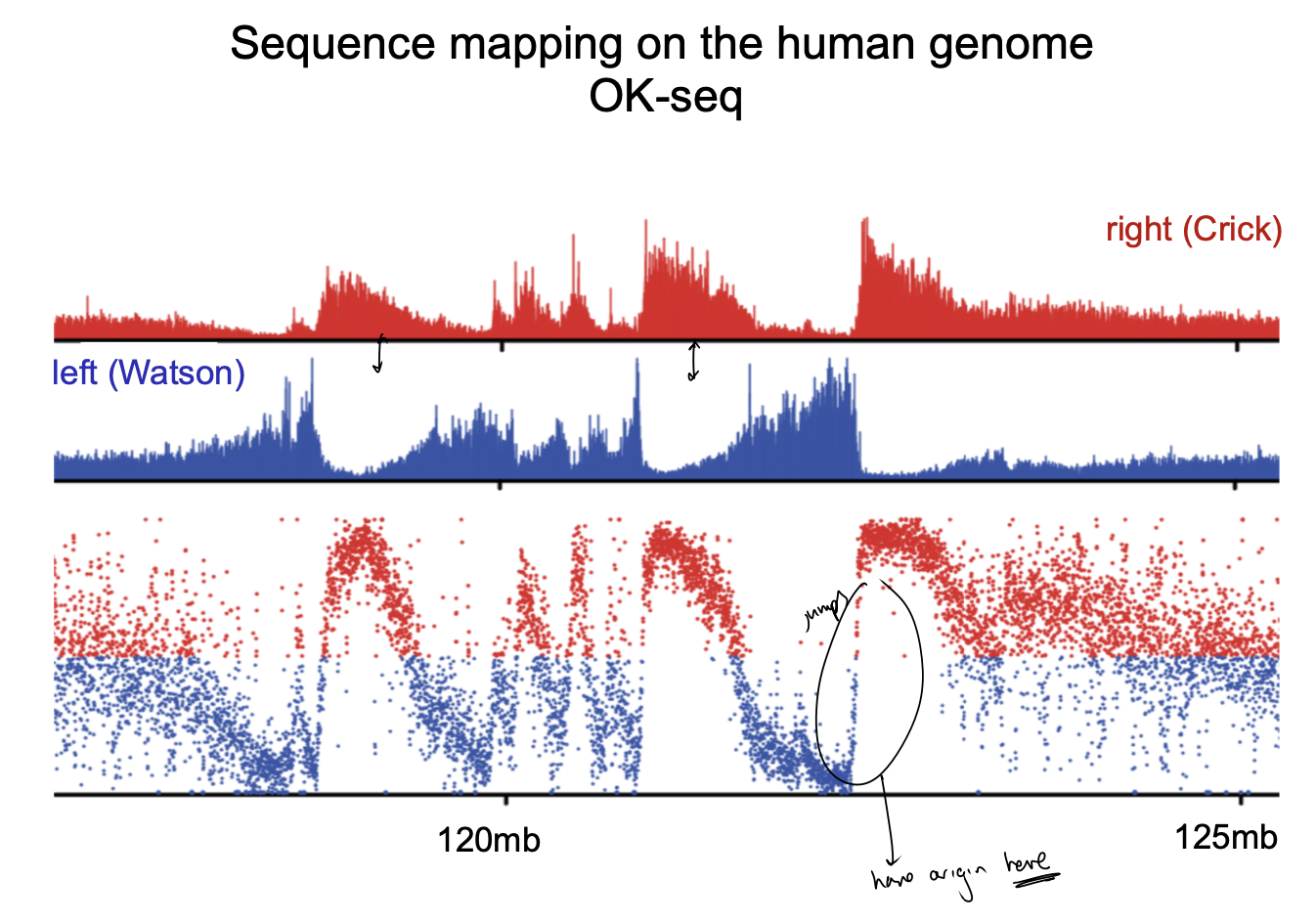
Sequence mapping on the human genome OK-seq
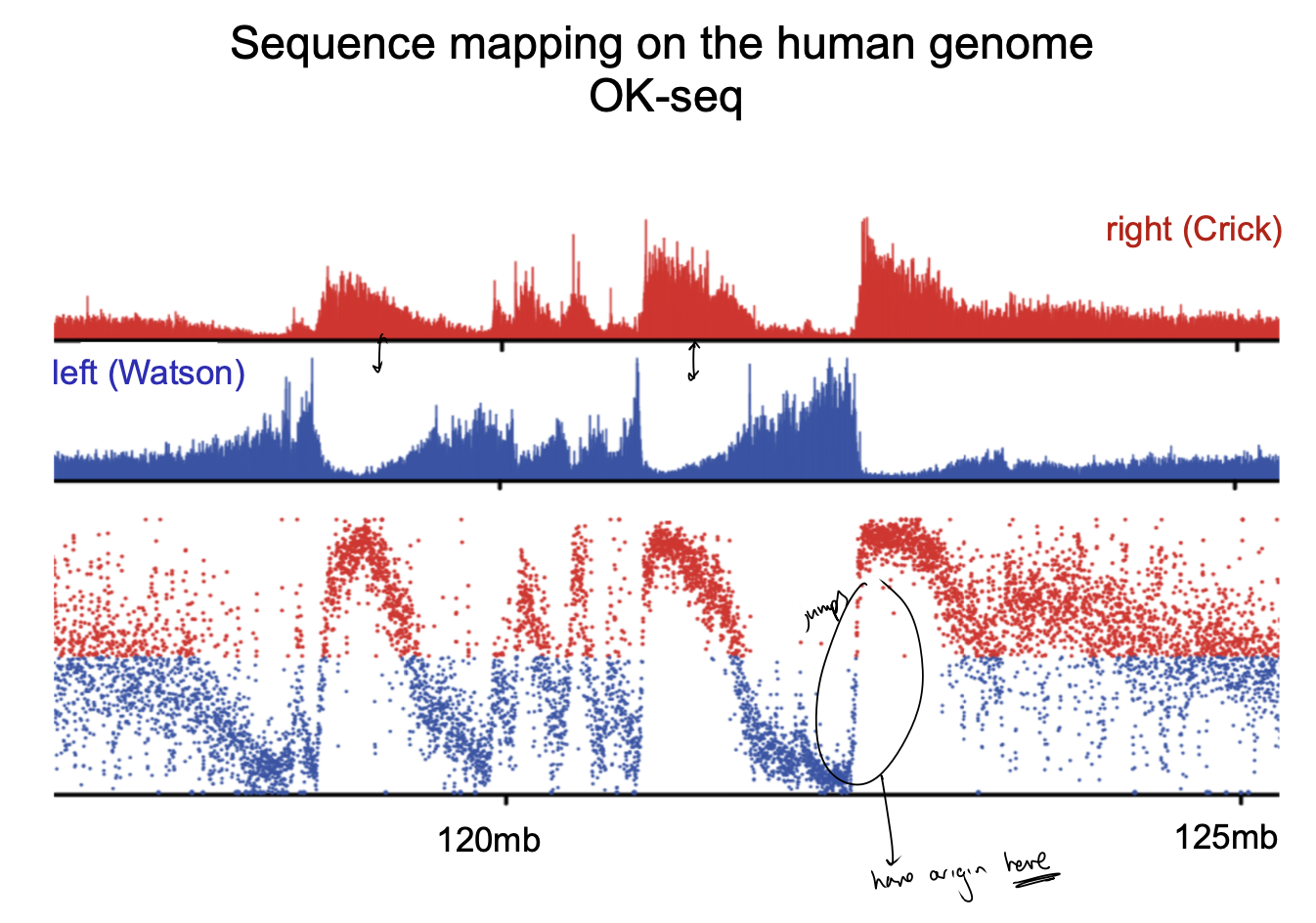
Sequence mapping on the human genome OK-seq compared to the other techniques: what does it show?
Initiation zone:
many inefficient origins
often flanked by efficient ones
→ conglomerate of many weak origins
What characterises active DNA replication origins?
not defined by consensus DNA sequences
unlike seen in yeast or SV40
What are these replication origin correlate to?
Proximity to gene promoter sites
but outside the genes
genomic sites with high GC content
including CpG islands
G quadruplexes
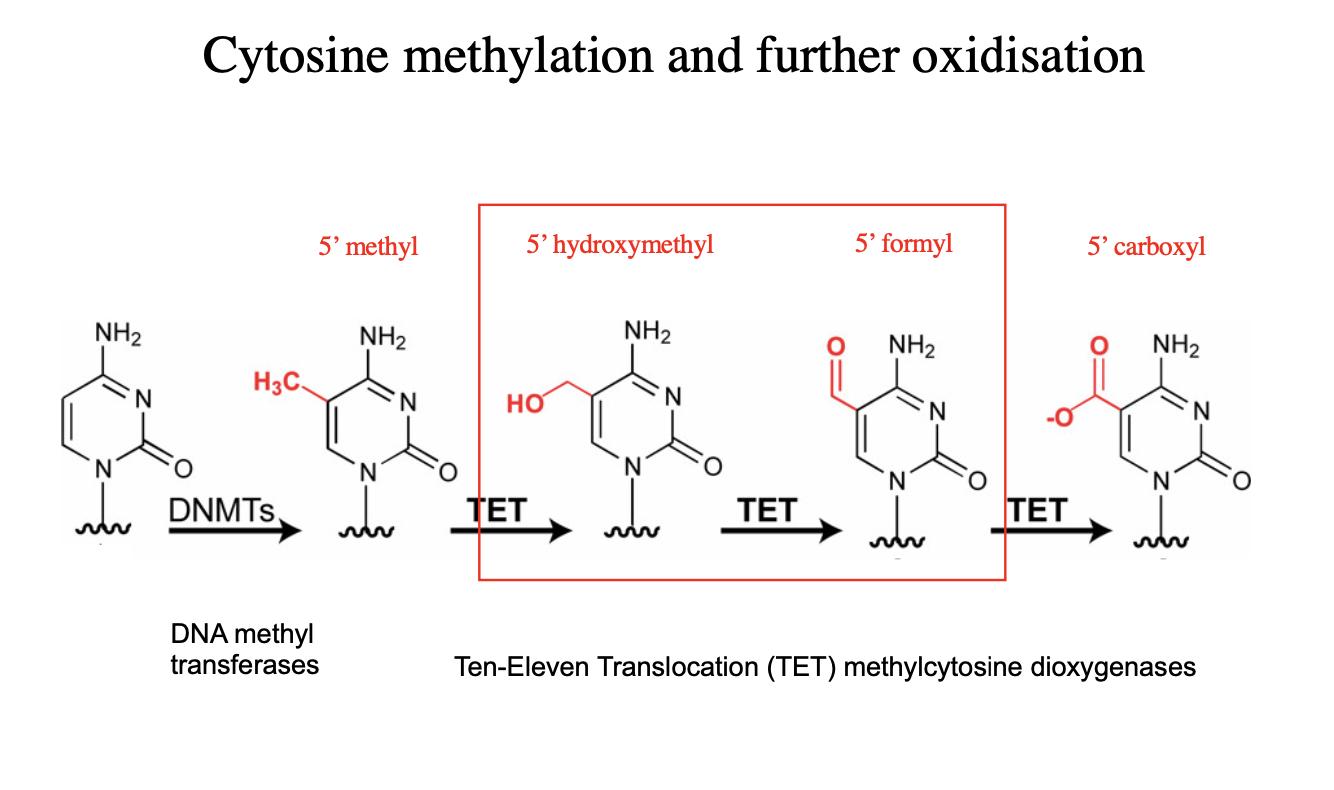
oxidised methyl-cytosines
Distinct patterns of histone modification
Open chromatin (hypersensitive to DNAse I)
Many of them!
Features of the genomic sites with high GC content
May form G quadruplex strucutres
May contain modified bases
methylated
hydroxymethylated cytosines
Additional experimental approaches
genome-wide profiling of Osaki fragments
isolation of small replication bubbles
These techniques have suggested that…
several individual origins tend to aggregate into
larger initiation zones
Prominent origin sites (and initiation zones) are often present in…
actively transcibed euchromatic regions of the genome
Where are they frequenctly located
at or upstream of Transciption Start Sites
What does this mean?
when DNA replication initiates at or near active gene promoters
replication and transciption elognation complexes move in the same direction
and any detrimental head-on collisions between are avoided
over that transcribed gene body!
Genetic and epigeneic factors that determine or influence specification of an origin site on a higher eukaryotic are…
still unclear and currently under investigation
Unlike in yeast…
No strict DNA consensus motifs are known (to date)
however
likely candidates of origin specification elements include:
short GC-rich DNA sequence motifs
that may lead to unusal DNA strucutures (i.e G quadruplexes)
include specific DNA and histone modifications
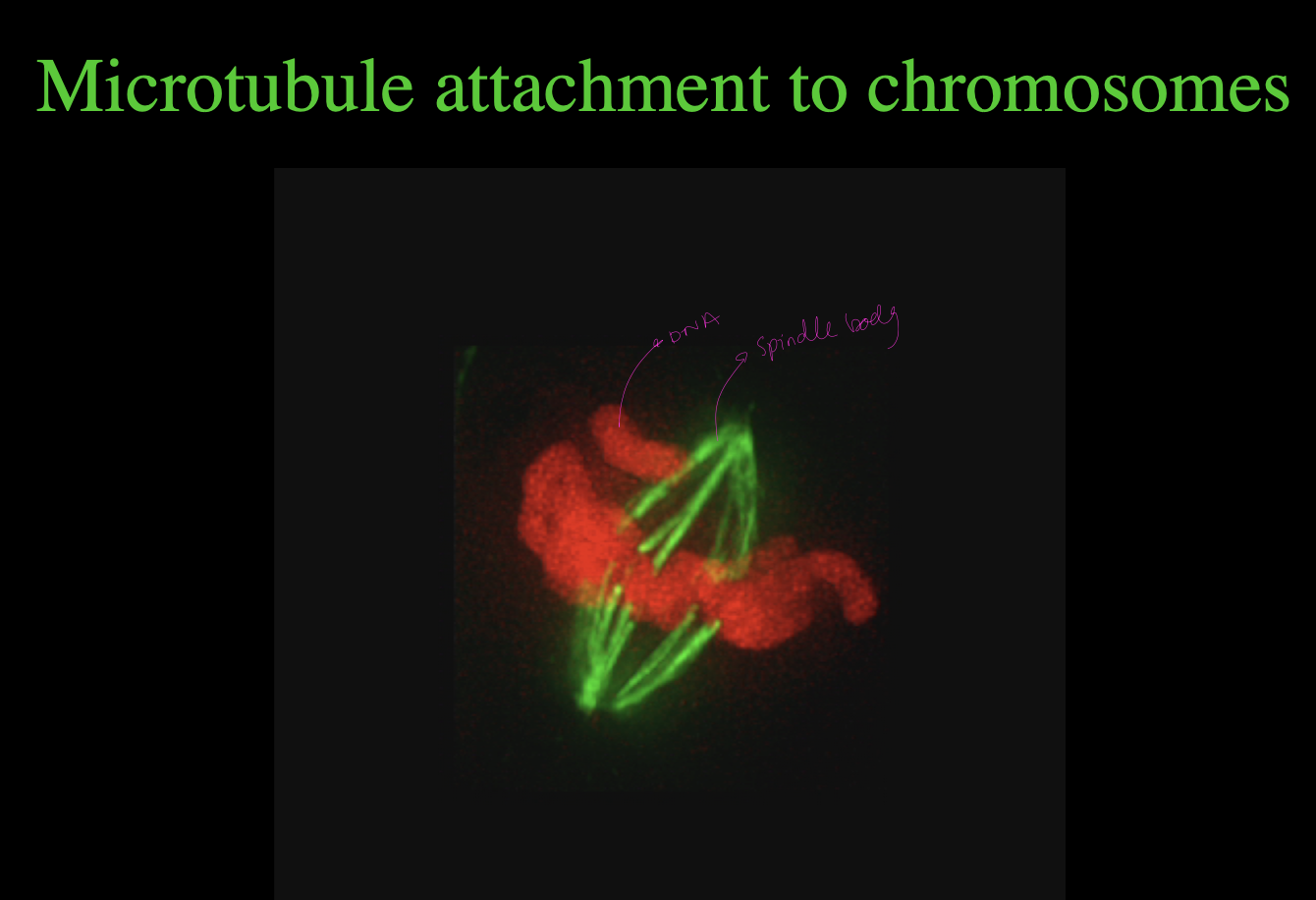
Centromeres: what do they do
attach chromosomes to mitotic or meiotic spindles
In yeast: why are they needed
Plasmids containing ARS elements replicate
BUT
are gradually lost without strong selective pressure
Centromere sequence (CEN)
stabilise them
We only x2 the DNA
so in order to enusre that each new cells has a copy of each chromosome→ need to make sure the DNA is exactly split in two
so need centromeres
unlike to bacteria which could just replicate loads of times and then hope that each cell will have a copy o each gene it needs.
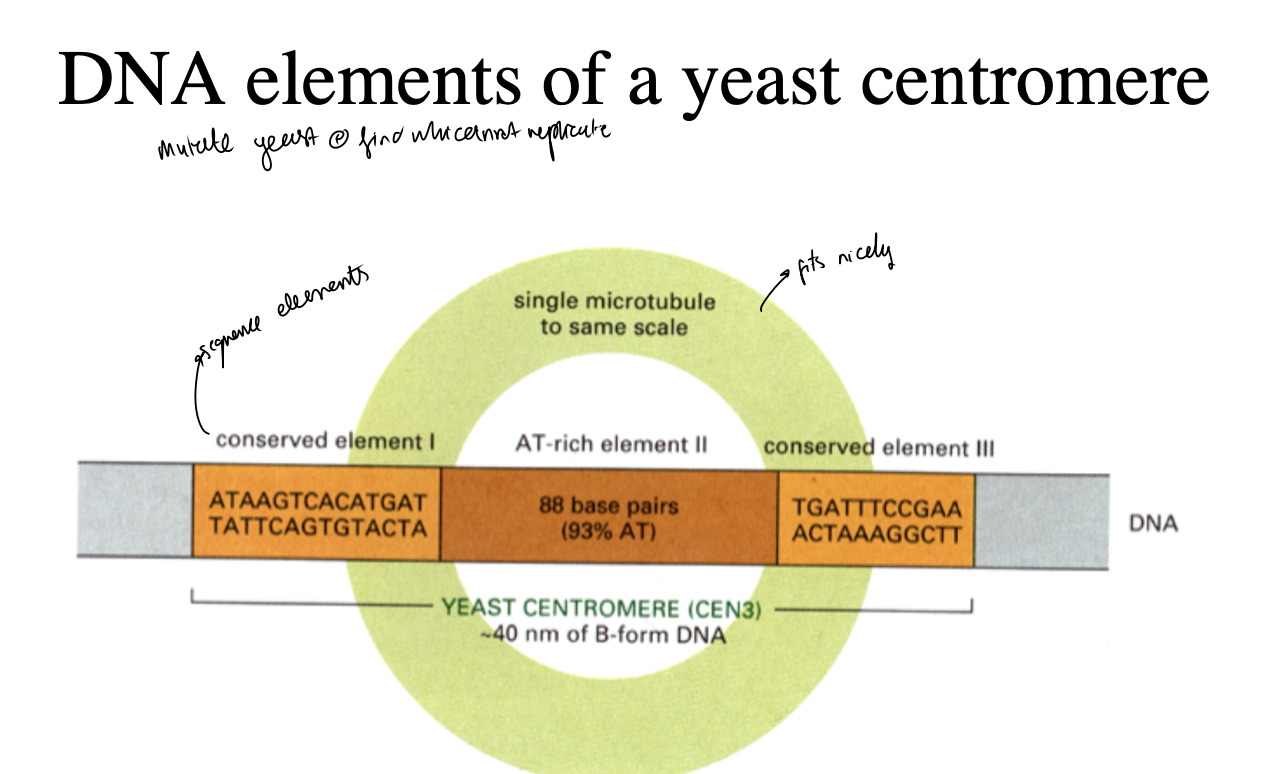
A CEN conist of
Three conserved DNA sequence elements:
I, II and III
Needed for SPECIFIC attatchment points for microtubule
AT-rich element n the middle
just needs to be the right size
doesn’t matter about sequence
so the microtubule can fit between the points
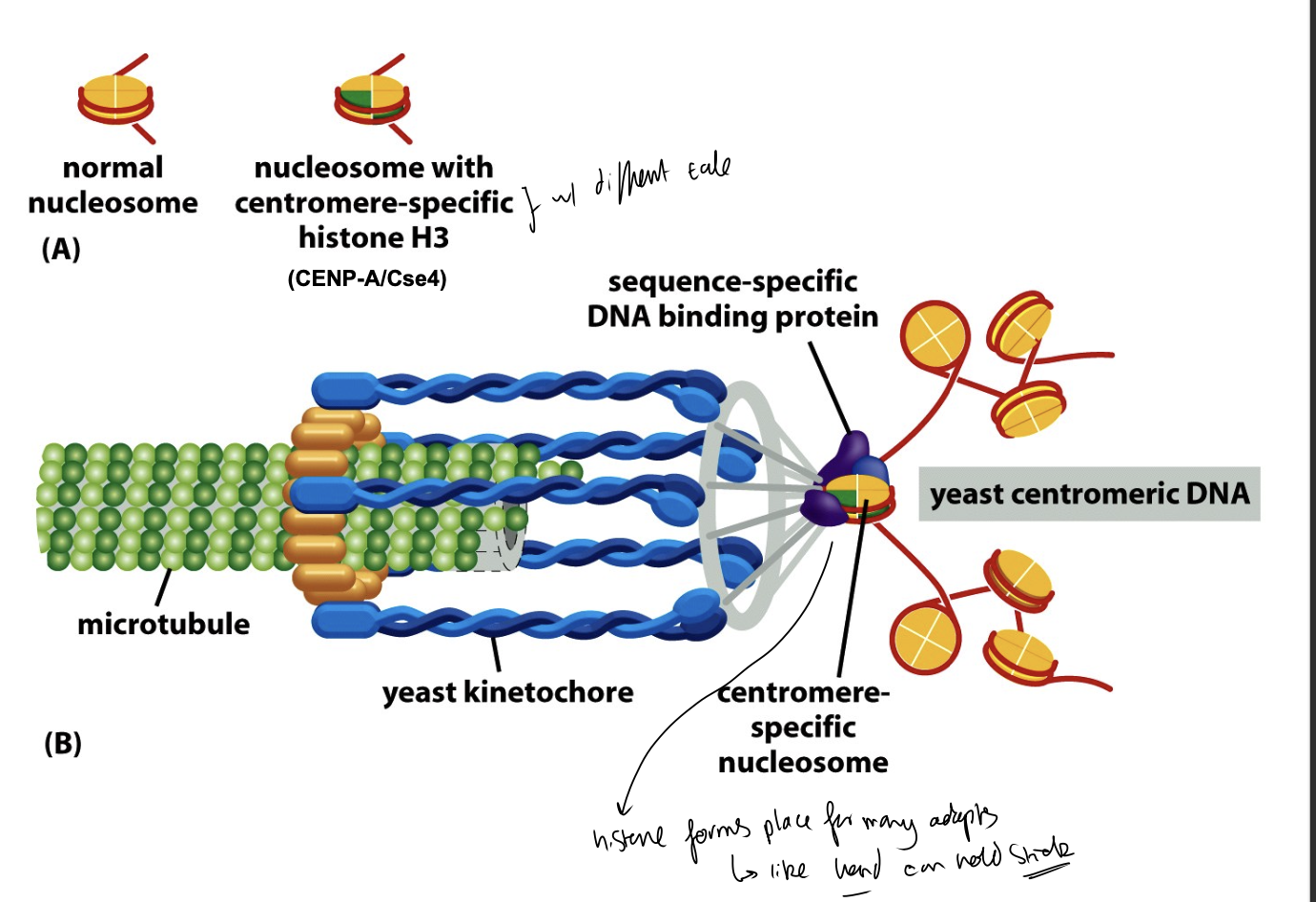
Centromes serve as what?
the attachment sites for
centromeric proteins
spindle microtubules
to form part of…
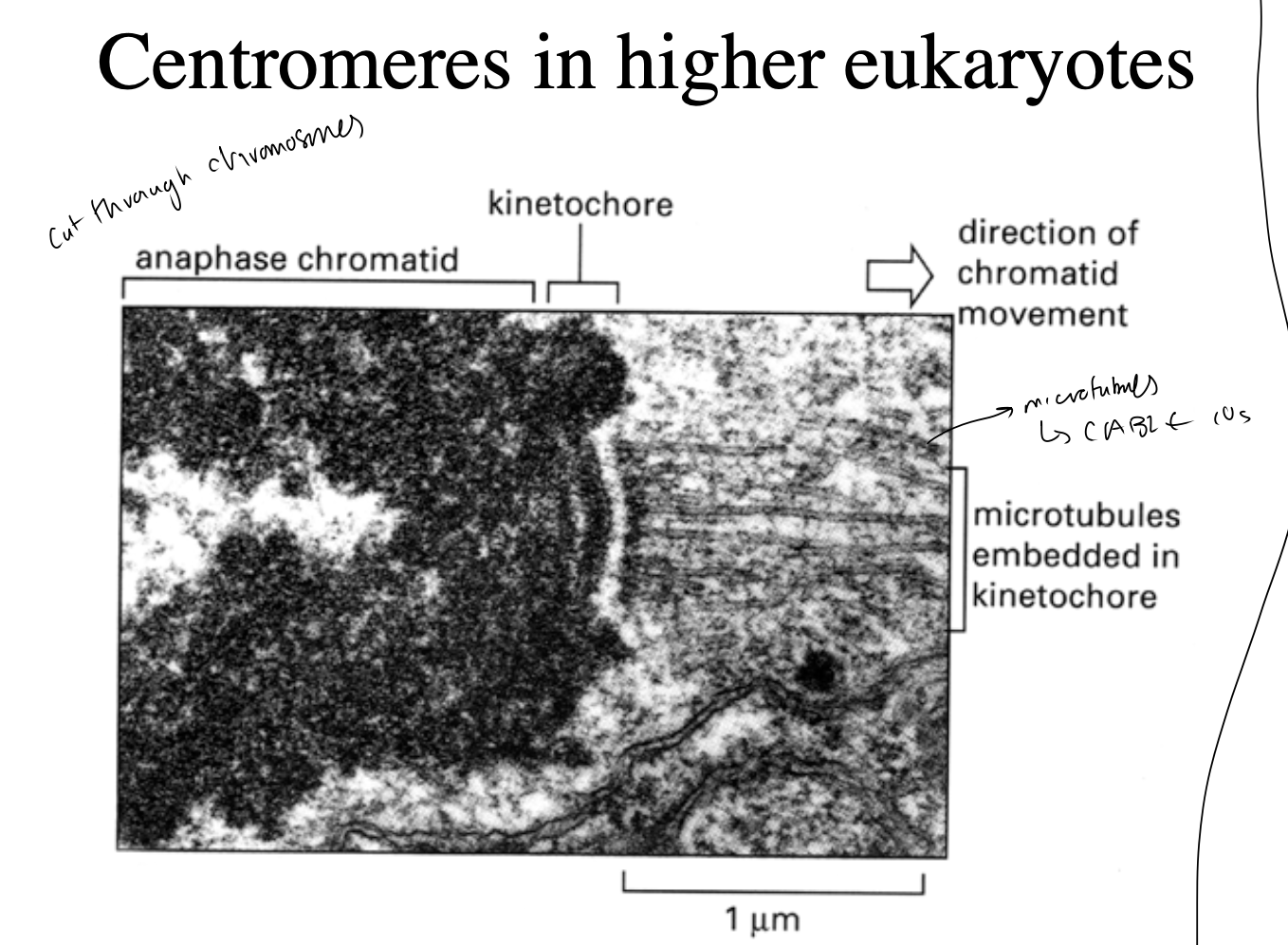
the kinetochore complex
where in mitosis, the chromatids of condensed chromosomes are attached to mitotic spindle
i.e microtubule does not attach directly→ instead does it through kinetochores
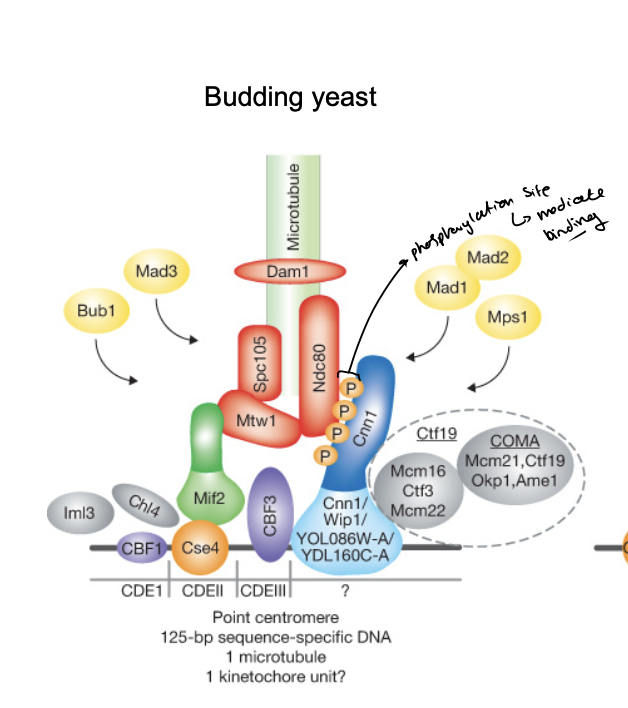
Molecular interactions at kinetochores
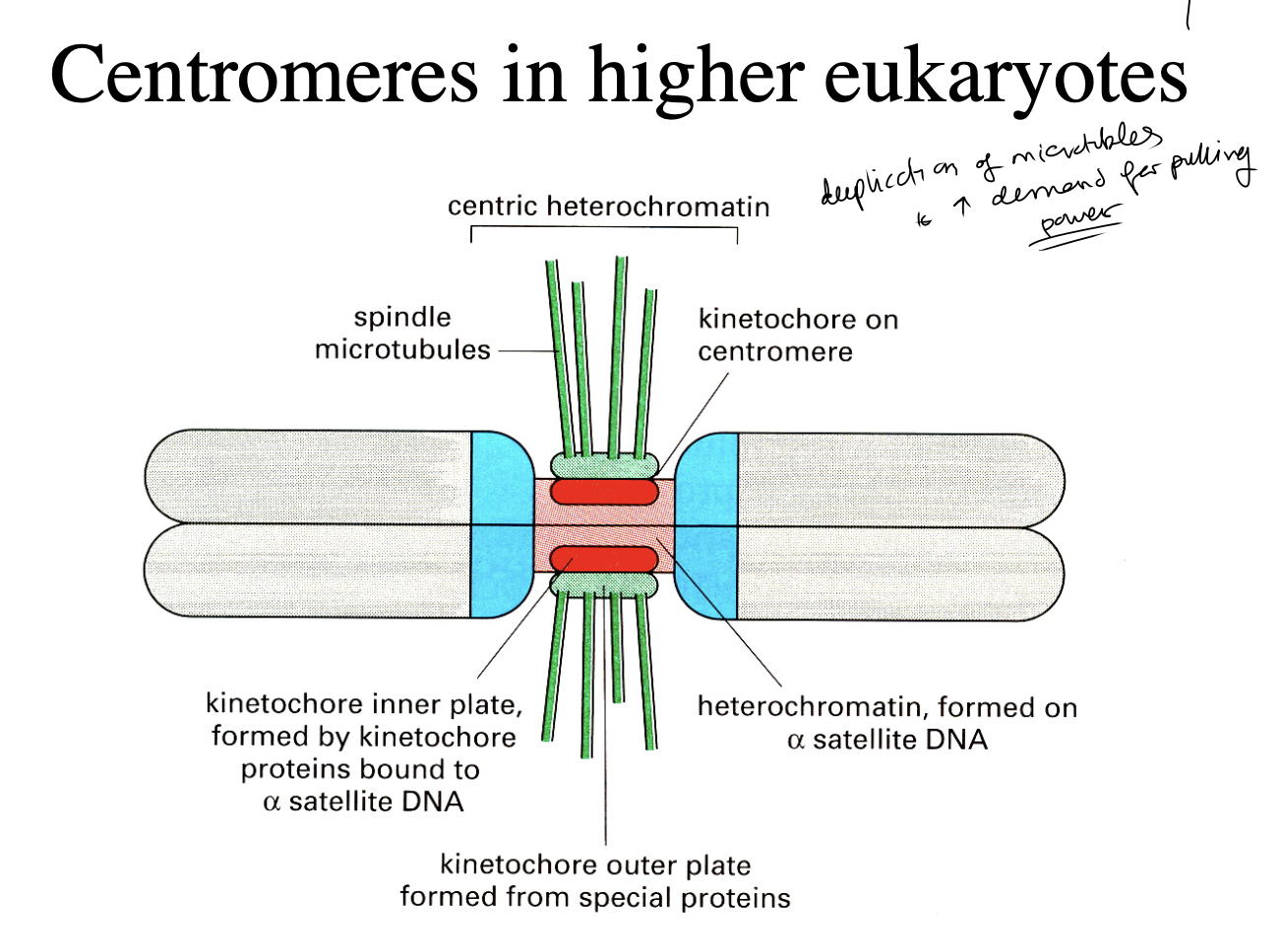
Centromeres in higher eukaryotes
Larger and more complex
you can see that they are similarin strucutre to the yeast ones
when interacting with other proteins
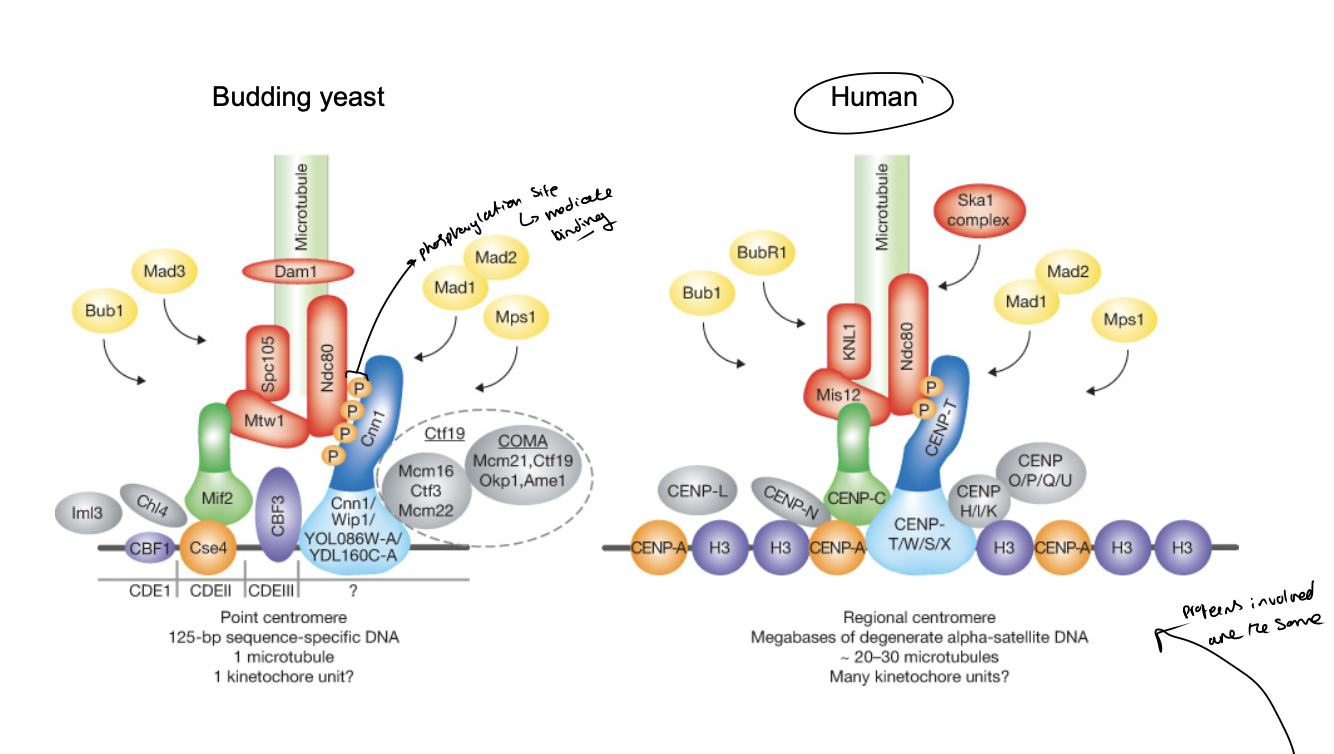
Centromeric DNA contains…
alpha-satellite DNA elements
further assembled into specific centromeric heterochomatin
spreads even further into chromosomes
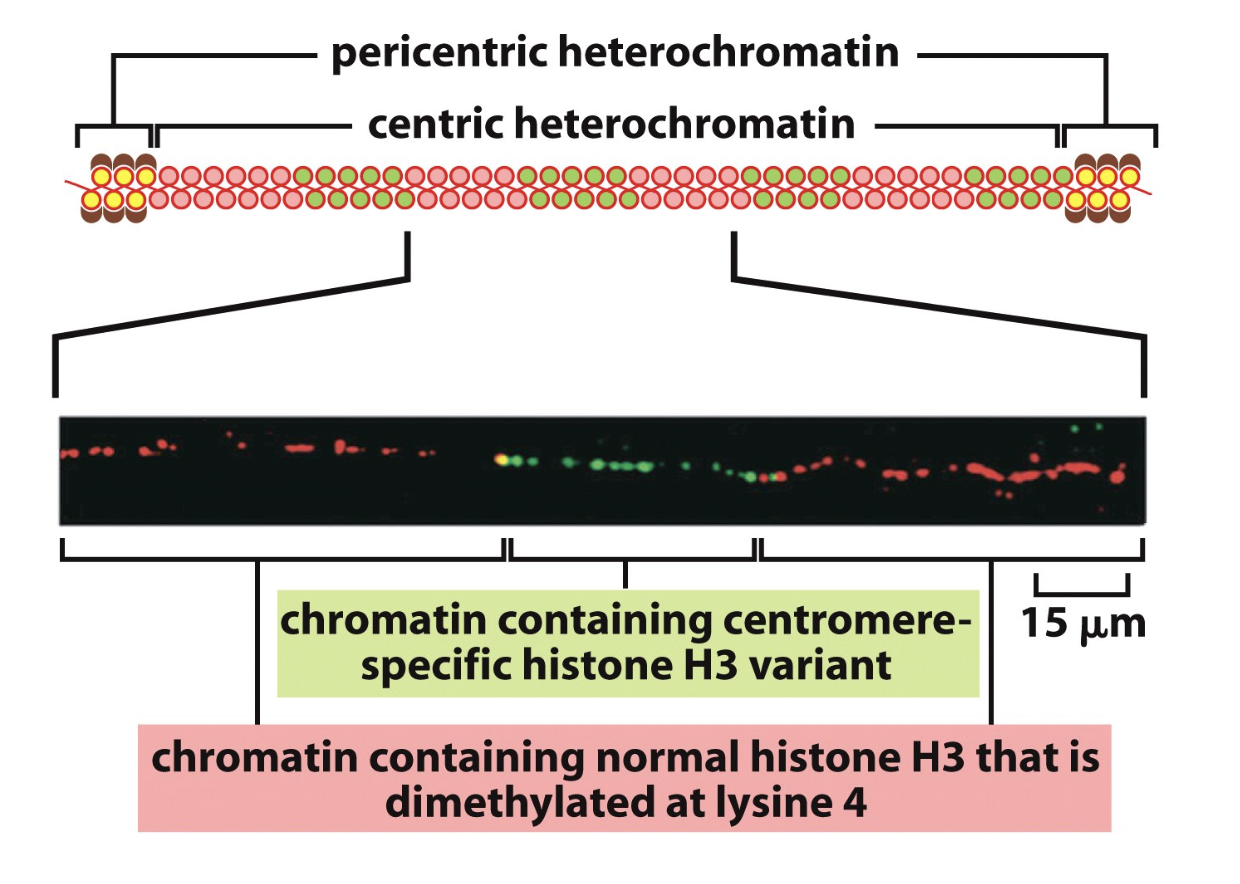
Centric heterochromatin contains…
sections with centromere-specific histone H3 varianet
sections with normal histone H3 that is di-methylated at lysine 4
Where centromeres are in the chromatin
What do telomeres provide
stable chromosome ends
Why need Telomeres
Protect from exonuclease degredation of ends
Stops the termination problem issue
when okaski fragments cannot be extended until the end
so causes degredation
telomeres→ before useful DNA so that it is only rubbish repeated regions that get degraded
telomerase can extend these in cells that do not really have a lifespan: Germ cells, cancer cells and induced in tissue culture cells
Linear plasmids in yeast need
CEN
ARS
Telomere (TEL)→ functional chromosome end
in order to replicate and segregate to daughter cells
Telomere DNA sequence repeats
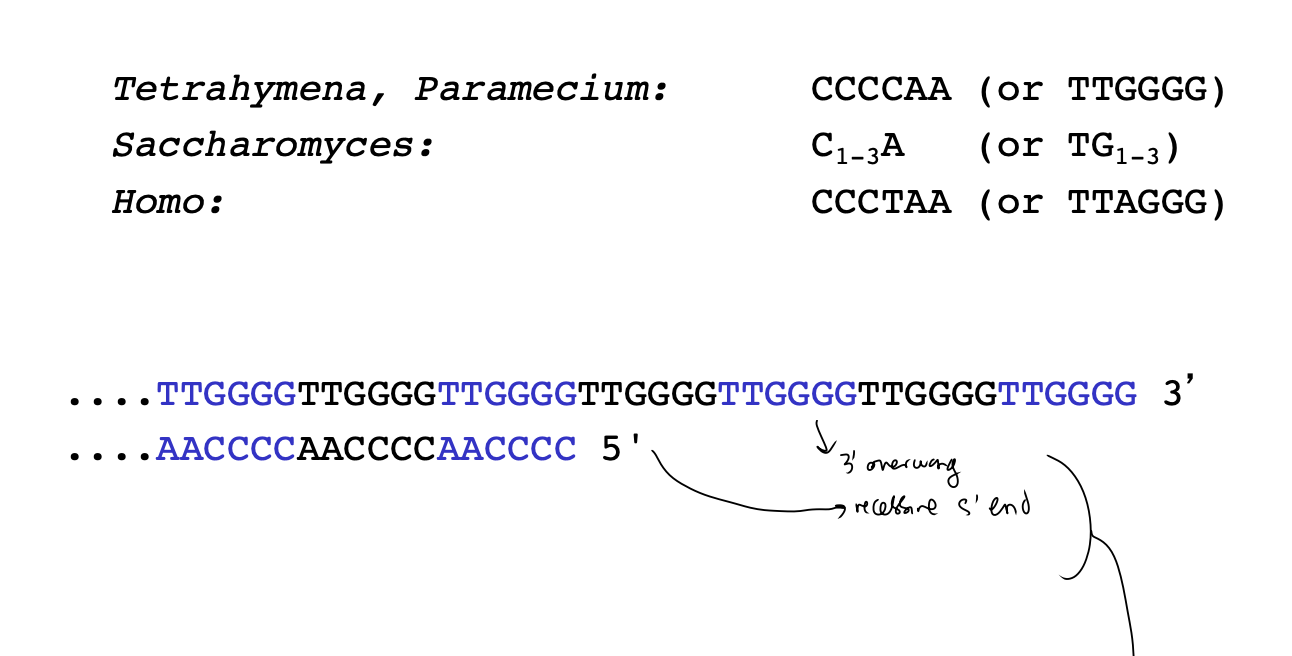
telomeres contain…
simple repeating sequences
forming 3’-single stranded DNA overhang ends
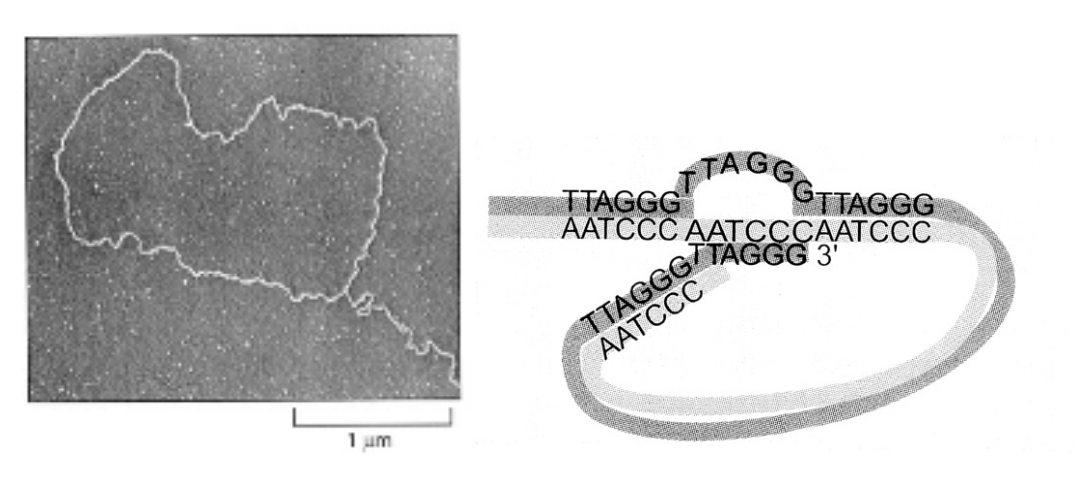
→ FORMS into Terminal loop T-loop
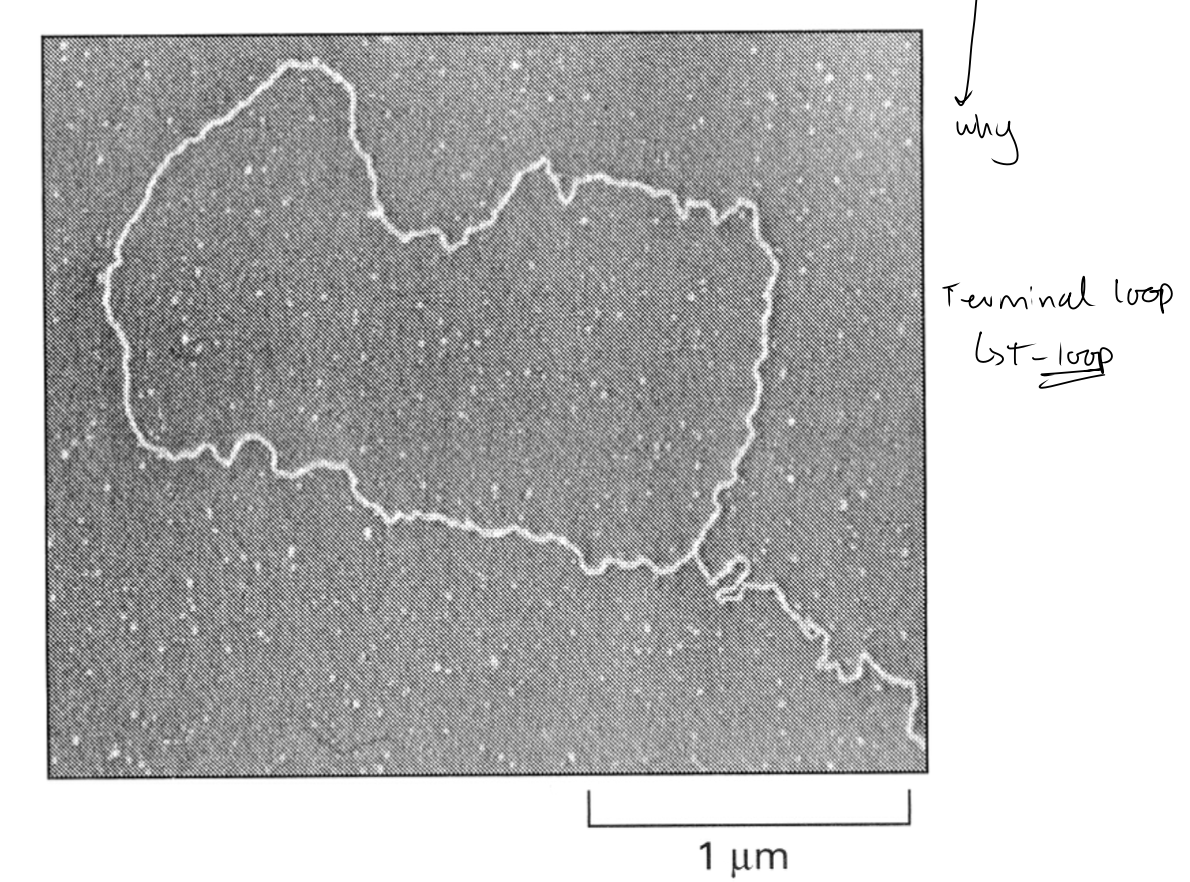
model of T-loop
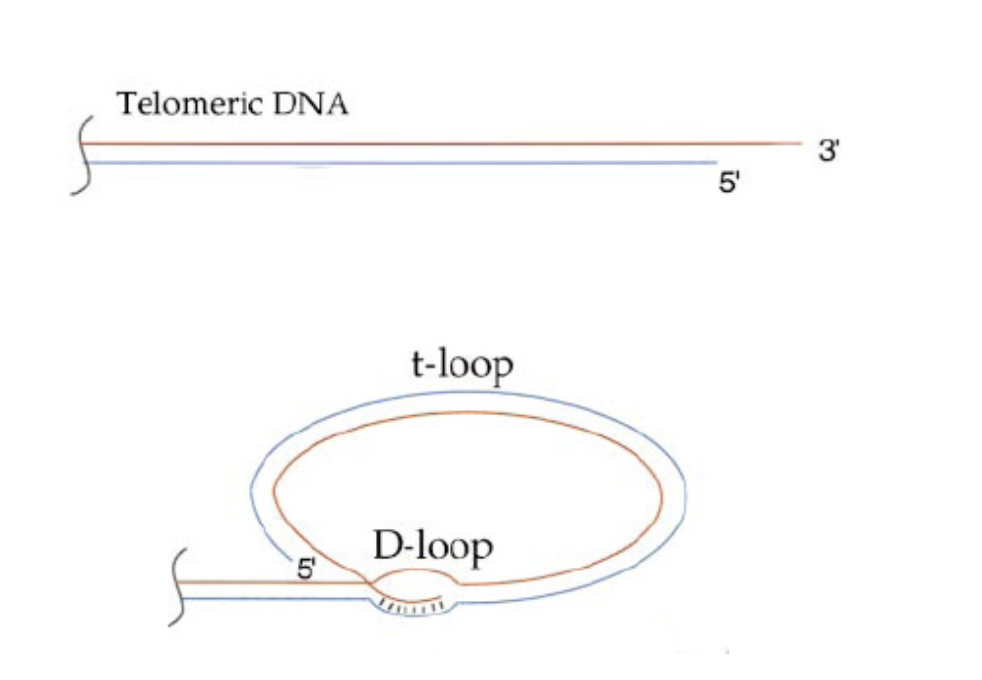
What are these 3’-single stranded DNA overhanging ends in different things
Yeast repeat TG(1-3)
human repeat TTAGG
Arabidopsis repeat TTTAGGG
stabilised by proteins
→ becomes a loop so no end anymore!
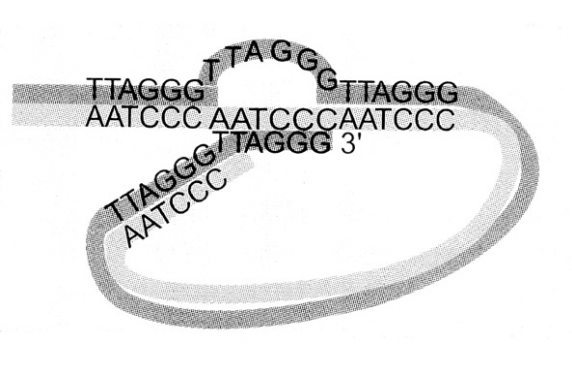
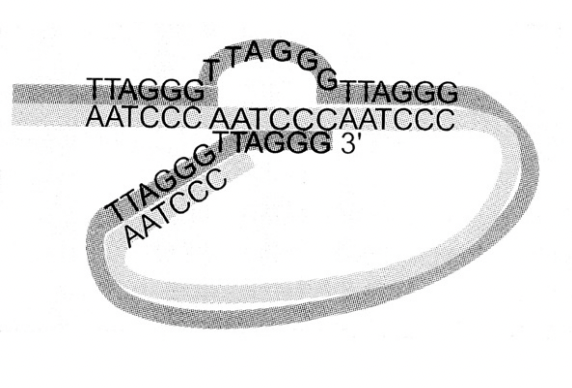
Models of the unsual strucutre of the DNA ends
hairpin
loop model
To explain the DNA ends as no free ends are detectable
EM has shown human telomere terminal loop ‘T-loop’
How was telomeric DNA assembled into
specilaised telomeric heterochromatin
attracts a protective protein complex called→ shelterin
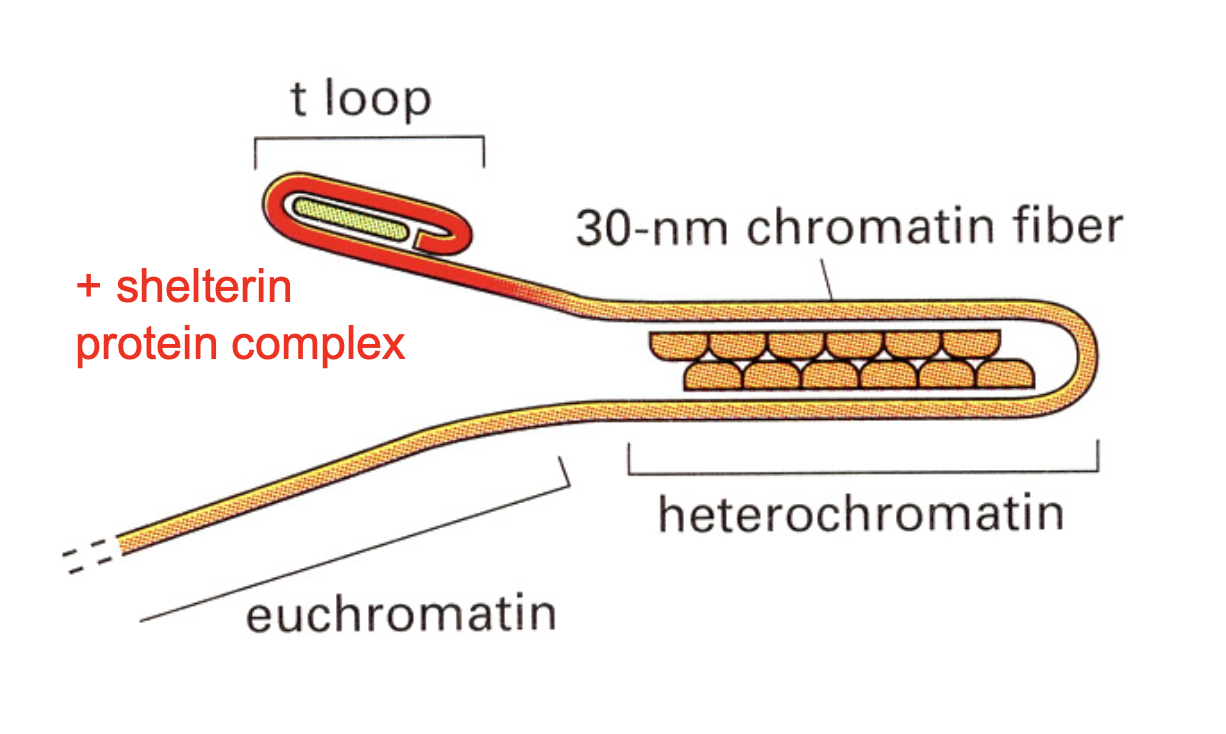
Telomeric heterochromatin spreads from…
telomeric DNA
further into the chromosomal DNA
It is possible to generate…
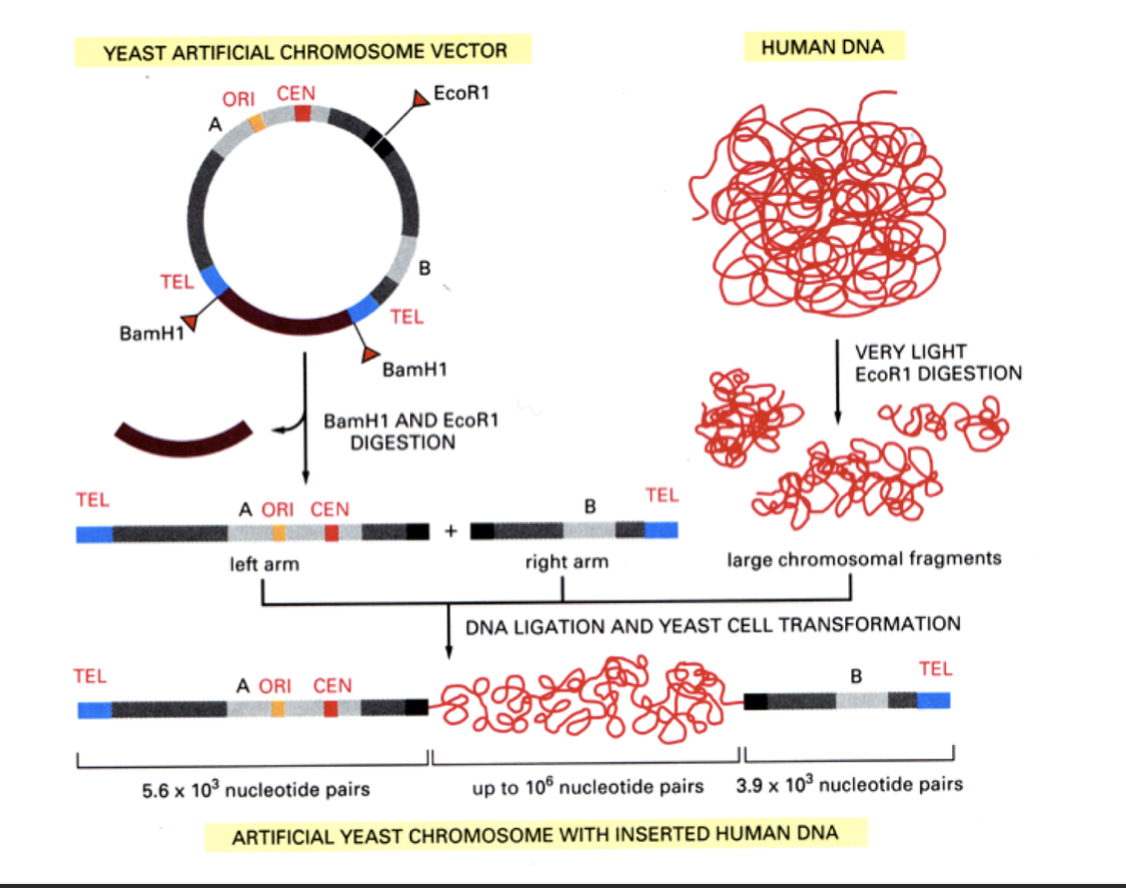
YACs→ yeast artificial chromosomes by using
all three elements
What are YACs used for
cloning huge DNA fragments
e.g
during the original human genome sequencing project
Hman sticky ends that can stick plasmid→ can transcribe useful human genes and get alot of DNA into it (unlike in bacteria genes)